Featured
-
-
Letter |
 Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection
Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection
The transcription cofactor CBF-β is shown to regulate the ability of HIV-1 to evade host restriction mediated by the deaminase APOBEC3; it acts by interacting with the HIV protein Vif, so disrupting the Vif–CBF-β interaction could provide a new therapeutic target against HIV-1 infection.
- Stefanie Jäger
- , Dong Young Kim
- & Nevan J. Krogan
-
Brief Communications Arising |
 GSK-3α/β kinases and amyloid production in vivo
GSK-3α/β kinases and amyloid production in vivo
- Tomasz Jaworski
- , Ilse Dewachter
- & Fred Van Leuven
-
Letter |
 GlcNAcylation of histone H2B facilitates its monoubiquitination
GlcNAcylation of histone H2B facilitates its monoubiquitination
Genome-wide analysis shows that H2B S112 O-linked to N-acetylglucosamine is frequently located near transcribed genes, suggesting that histone GlcNAcylation facilitates transcription of the genes.
- Ryoji Fujiki
- , Waka Hashiba
- & Shigeaki Kato
-
Letter |
 Control of Drosophila endocycles by E2F and CRL4CDT2
Control of Drosophila endocycles by E2F and CRL4CDT2
- Norman Zielke
- , Kerry J. Kim
- & Bruce A. Edgar
-
Letter |
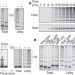 Protein targeting and degradation are coupled for elimination of mislocalized proteins
Protein targeting and degradation are coupled for elimination of mislocalized proteins
- Tara Hessa
- , Ajay Sharma
- & Ramanujan S. Hegde
-
Letter |
 UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids
UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids
- Dawn M. Wenzel
- , Alexei Lissounov
- & Rachel E. Klevit
-
Letter |
 Neuropsin cleaves EphB2 in the amygdala to control anxiety
Neuropsin cleaves EphB2 in the amygdala to control anxiety
- Benjamin K. Attwood
- , Julie-Myrtille Bourgognon
- & Robert Pawlak
-
Letter |
 SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis
SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis
The ubiquitin conjugation system regulates the canonical NF-κB activation pathway, which mediates immune responses. Linear polyubiquitin chains—in which the C terminal glycine of ubiquitin is conjugated to the α-amino group of the amino-terminal methionine of another ubiquitin—are generated by a unique ubiquitin ligase complex called linear ubiquitin chain assembly complex (LUBAC) composed of two RING domain proteins called HOIL-1 and HOIP. This is one of three complementary studies identifying a novel component of the LUBAC complex called SHARPIN, which is recruited to receptor signalling complexes (RSCs) that form after TNF and CD40L stimulation. The LUBAC complex containing SHARPIN stimulates the formation of linear ubiquitin chains in vitro and in vivo and is required for the activation of NF-κB signalling.
- Fumiyo Ikeda
- , Yonathan Lissanu Deribe
- & Ivan Dikic
-
Article |
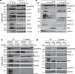 Linear ubiquitination prevents inflammation and regulates immune signalling
Linear ubiquitination prevents inflammation and regulates immune signalling
- Björn Gerlach
- , Stefanie M. Cordier
- & Henning Walczak
-
Article |
 Structure and mechanism of the hexameric MecA–ClpC molecular machine
Structure and mechanism of the hexameric MecA–ClpC molecular machine
Regulated proteolysis by ATP-dependent proteases have a crucial role in protein quality control in cells. The Clp/Hsp100 proteins of the AAA+ superfamily of ATP-dependent chaperones unfold and translocate proteins into the proteolytic chamber of protease complexes. ClpC requires the adaptor protein MecA for activation and substrate targetting to the ClpCP protease complex. Here, a structural and biochemical analysis is presented of the MecA–ClpC complex revealing organizational principles and providing mechanistic insights into this complex molecular machine.
- Feng Wang
- , Ziqing Mei
- & Yigong Shi
-
News & Views |
 A new look for the APC
A new look for the APC
Solving the structure of protein complexes is particularly challenging when they contain many subunits. In the case of the APC, a fruitful strategy has been to gain information by subtracting subunits. See Article p.227 and Letter p.274
- Ian Foe
- & David Toczyski
-
Letter |
 Structures of APC/CCdh1 with substrates identify Cdh1 and Apc10 as the D-box co-receptor
Structures of APC/CCdh1 with substrates identify Cdh1 and Apc10 as the D-box co-receptor
The anaphase promoting complex/cyclosome (APC/C) is a large multimeric ubiquitin E3 ligase that regulates the eukaryotic cell cycle in processes such as chromatid segregation and completion of mitosis. It catalyses the polyubiquitylation of a diverse array of mitotic regulatory proteins and targets them for proteasomal degradation. Target selection also involves a co-activator protein (either Cdc20 or Cdh1) together with core APC/C subunits. Here, a cryo-EM structure of APC/CChd1 bound to a D-box peptide substrate is presented. The structure provides important insight into the recognition and catalytic mechanism of APC/C substrates.
- Paula C. A. da Fonseca
- , Eric H. Kong
- & David Barford
-
Letter |
 The proteasome antechamber maintains substrates in an unfolded state
The proteasome antechamber maintains substrates in an unfolded state
The proteasome is a multi-protein complex that enzymatically degrades proteins. Proteolysis occurs in a barrel-shaped 20S core particle comprising three interconnected cavities, including a pair of antechambers in which substrates are held before degradation. These authors demonstrate that substrates interact actively with the antechamber walls and that the environment in this compartment is optimized to maintain the substrates in unfolded states so as to be accessible for hydrolysis.
- Amy M. Ruschak
- , Tomasz L. Religa
- & Lewis E. Kay
-
Letter |
 Role of a ribosome-associated E3 ubiquitin ligase in protein quality control
Role of a ribosome-associated E3 ubiquitin ligase in protein quality control
The translation of messenger RNA that lacks stop codons results in the production of aberrant proteins, which may have harmful effects on the cell. It is unclear how eukaryotic cells eliminate these 'non-stop' proteins. Here it is shown that, in Saccharomyces cerevisiae, an E3 ubiquitin ligase called Ltn1 acts in the quality-control pathway. It associates with ribosomes and marks non-stop proteins with ubiquitin, which targets the proteins for degradation.
- Mario H. Bengtson
- & Claudio A. P. Joazeiro
-
Article |
 Enhancement of proteasome activity by a small-molecule inhibitor of USP14
Enhancement of proteasome activity by a small-molecule inhibitor of USP14
In the ubiquitin–proteasome system, substrates destined for destruction are modified with ubiquitin chains and then degraded by the proteasome. These authors reveal a regulatory mechanism in which proteasomal activity is modulated by the length of ubiquitin chains in human cells. They find that deubiquitinating enzyme USP14 can inhibit the degradation of ubiquitin-conjugated substrates by trimming ubiquitin chains, and that stimulation of proteasome activity may be used to reduce the levels of toxic proteins in cells.
- Byung-Hoon Lee
- , Min Jae Lee
- & Daniel Finley
-
Article |
 Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii
Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii
Although Archaea encode proteasomes highly related to those of eukaryotes, archaeal ubiquitin-like proteins are less conserved and not known to function in protein conjugation, complicating our understanding of the origins of ubiquitination. Two small archaeal modifier proteins, SAMP1 and SAMP2, structurally similar to ubiquitin, are now reported to form protein conjugates in the archaeon Haloferax volcanii.
- Matthew A. Humbard
- , Hugo V. Miranda
- & Julie A. Maupin-Furlow

 FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas
FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas