Abstract
The genetic influence on human vocal pitch in tonal and non-tonal languages remains largely unknown. In tonal languages, such as Mandarin Chinese, pitch changes differentiate word meanings, whereas in non-tonal languages, such as Icelandic, pitch is used to convey intonation. We addressed this question by searching for genetic associations with interindividual variation in median pitch in a Chinese major depression case-control cohort and compared our results with a genome-wide association study from Iceland. The same genetic variant, rs11046212-T in an intron of the ABCC9 gene, was one of the most strongly associated loci with median pitch in both samples. Our meta-analysis revealed four genome-wide significant hits, including two novel associations. The discovery of genetic variants influencing vocal pitch across both tonal and non-tonal languages suggests the possibility of a common genetic contribution to the human vocal system shared in two distinct populations with languages that differ in tonality (Icelandic and Mandarin).
Similar content being viewed by others
Introduction
Human speech production is a complex process involving not only the coordinated activity of various organs1, but also the social and cultural environments in which people learn to speak. As with all complex physiological processes, genetic effects likely play a role, but their extent and their molecular basis are largely unknown. Most prior molecular genetic studies have focused on disorders of language in a broader sense, such as disorders of reading and writing and developmental speech and language impairments2,3, work that led to the identification of mutations in single genes that cause speech disorders4. Far fewer studies have looked at the genetic basis of speech production with acoustic measures5, and it is unclear to what extent, if any, phonation characteristics are learnt rather than inherited (or influenced by the interplay of both). Finding the molecular basis of speech acoustics could shed light on this key, uniquely human, attribute.
There is a broad set of acoustic measures, each representing a specific aspect of the human vocal system and its psychological correlates6. A recent genome-wide association study (GWAS) discovered the first genetic locus associated with median voice pitch6, which reflects the rate of vocal fold vibration and is perceived as how deep or high the voice sounds. By recording the voice of 12,901 Icelanders during a reading task, a single locus was found that exceeded a corrected significance threshold, on chromosome 12 at the adenosine triphosphate binding cassette subfamily C member 9 gene (ABCC9).
The Icelandic finding, so far not replicated in an independent sample, raises several questions: First, does the chromosome 12 locus have the same effect in other populations? Iceland is genetically and linguistically homogeneous with minor dialectal variation in their non-tonal language6. Second, how does the chromosome 12 locus affect pitch in speakers of tonal languages? Pitch often represents word emphasis and speakers’ emotional context and can convey semantic information, especially in tonal (or pitch-accented) languages, where pitch is used to differentiate word meanings7. Around 60–70% of the world’s languages are tonal languages7. Third, does the effect exist in spontaneous speech, or is it restricted to a reading task, like the one used in the Icelandic study? Finally, given reported differences in pitch attributable to variation in mood8,9,10, to what extent do the findings depend on the mood of the subjects? To address these questions, we performed GWAS on voice pitch measured in 7654 Han Chinese Women, recruited for a case-control genetic study of the origins of major depressive disorder (MDD). The design allowed us to incorporate changes in mood into our analysis and to explore whether genetic effects on pitch were the same or different in tonal and non-tonal languages.
Results
About 364,929 voice segments were manually identified from 7654 subjects (3641 cases and 4013 controls). The mean duration of audio extracted from case interviews was 192.73 s (SD = 196.68), approximately twice as long as for controls (97.44 s, SD = 122.96). Segments were manually classified for their noise level and accent. 60% of the subjects spoke in standard Mandarin, whereas the rest spoke either their local languages or Mandarin with local accents. 78% of the interviews were recorded with no or low noise levels. The voice and demographic information in the case/control subgroups are reported in Supplementary Table 1.
We used the median F0 to measure pitch, the same measure used in the Iceland study6. The mean over all subjects was 206.25 Hz (SD = 26.31 Hz). Pitch was associated with age (\({{{{{\rm{\beta }}}}}}=-0.25,{P}=9.65\times {10}^{-108}\)), and, after correcting for the effects of the collection site, weakly associated with MDD \({{{{{\rm{\beta }}}}}}\) = 0.14, P = 0.013). After adjusting for age and MDD, pitch was significantly associated with height, BMI, education level, and other confounding variables listed in Supplementary Table 2.
The heritability of pitch was 20% (95% CI: 10 to 29%, close to the estimation in the Iceland study: 17%, 95%CI: 9 to 24%) after adjusting for MDD and other covariates as listed in Supplementary Table 2. The genetic correlation of pitch between MDD cases and controls was not significantly different from 1 (rg = 1.00, SE = 0.43, P = 0.5). However, we conservatively performed GWAS on the two cases and controls separately and then combined the effects using meta-analysis. No genome-wide significant loci were identified (Fig. 1). The variant rs11046212-T in an intron of the ABCC9 gene, associated with a pitch in the Iceland study6, was one of the strongest associations in our study (\(\beta =0.09{SD}\), 2.3 Hz per allele, \(P=2.33\times {10}^{-7}\)). Association with one other locus on the same chromosome, rs10859172-C, achieved almost equal significance (\(\beta =-0.08{SD},{P}=2.06\times {10}^{-7}\)). We tested for heterogeneity and found that the effects of these two SNPs were not heterogeneous between cases and controls (rs11046212, heterogeneity P value = 0.61; rs10859172, heterogeneity P value = 0.83).
We explored the association with two other measures of pitch, the first and the third quartile of F0 (see Supplementary Note 1, Supplementary Figs. 1–3, and Supplementary Table 3 for a comparison of different quantiles of F0.). Two loci, one associated with the first and one associated with the third quartile of F0, achieved genome-wide significance (Supplementary Table 4). They were rs11046212-T with the first quartile of F0 (\(\beta =0.10{SD}\), \(P=3.32\times {10}^{-8}\)), and rs10859172-C with the third quartile of F0 (\(\beta =-0.09{SD}\), \(P=2.53\times {10}^{-8}\)). Since we observed variation between hospitals in the distribution of voice features (Supplementary Table 5), we also ran a hospital-level meta-analysis and confirmed that the associations between SNPs and pitch were not heterogeneous between hospitals (Supplementary Note 2).
We compared several F0 statistics in our study with the 7278 females in the Iceland study. The results are shown in Supplementary Table 6. The pitch (F0 median) was significantly higher in the Chinese sample than in Iceland (t-statistic\(=8.31\), \(P=1.06\times {10}^{-16}\)). Pitch in our dataset varied more within-person than in the Iceland study (standard deviations of F0: t-statistic\(=56.17\), \(P < 1.23\times {10}^{-308}\)), and we observed higher skewness in the distribution of F0 in Chinese individuals (t-statistic \(=32.97\), \(P=3.14\times {10}^{-230}\)) which we attribute to the greater use pitch plays in conveying meaning in Chinese tonal languages than in Icelandic.
We then built a polygenic score (PGS) based on several different P value thresholds (PT) from the summary statistics provided by the Iceland study and tested the predictive performance of our data. The results are shown in Fig. 2. The best fit included only five SNPs at PT = \(2.30\times {10}^{-7}\), and explained 0.61% of the variance in pitch (P = \(3.32\times {10}^{-8}\)). The PRS model explained a proportion of variance in the case/control subgroups that was similar to that in the entire group. (Supplementary Figs. 4, 5).
We evaluated whether the observed fraction of results displaying the same direction of allelic effects across studies was significantly greater than expected by chance (that is, 50%) using binomial sign tests. Table 1 gives the number of LD-independent SNPs in the Iceland study at a set of P value thresholds, and the fraction of these SNPs displaying the same direction of effect in the Chinese group and a one-sided binomial test P value. 98.99% of the SNPs showed the same direction at a P value threshold <\(1\times {10}^{-6}\) (One-sided binomial P = \(1.58\times {10}^{-28}\)).
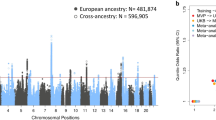
Finally, we performed a meta-GWAS combining the Iceland study and our case/control subgroups. The Manhattan plot is shown in Fig. 3. We found four genome-wide significant hits, of which two were novel. The four top hits from the cross-population meta-analysis, together with the rs10859172 found in our study, are listed in Table 2. The most significant association was for the rs11046212-T at the ABCC9 locus (P = \(7.50\times {10}^{-24}\)), which showed a similar effect in Chinese (β = 0.09 SD, 2.3 Hz per allele) and Icelanders (β = 0.11 SD, 2.1 Hz per allele). Its allele frequency, however, was lower in China (0.25) than in Iceland (0.48). Results of all SNPs that achieved genome-wide significance are in Supplementary Data 1.
a Manhattan plot of the cross-population meta-GWAS. The dashed horizontal line indicates genome-wide significance (top, \(5\times {10}^{-8}\)). Novel hits are marked in bold. b–e Regional plots of the top variants associated with voice pitch. The −log10(P value) of imputed SNPs associated with pitch is shown on the left y-axis. The recombination rates expressed in centimorgans (cM) per Mb (GRCh37; blue lines), are shown on the right y-axis. Position in Mb is on the x-axis. The plots were drawn using LocusZoom31.
Cross-ancestry fine-mapping analysis of the ABCC9 locus identified four variants as probable causal variants in the 95% credible set (Supplementary Table 7). The variant rs11046212 had the maximum posterior probability (0.35), which was inferred as causal in both EUR and EAS populations (population-specific causal probability >0.99).
Discussion
We were able to answer three questions about the genetic basis of pitch production. First, we established that at least some of the same genetic loci are present in both Chinese and Icelandic populations. The loci we identified contribute to voice pitch in both tonal and non-tonal languages. We have no evidence that the loci differ in direction or size of the effect, despite the marked differences in the structure of the languages and the semantic use of pitch in Mandarin. Second, the effects are found in both spontaneous speech as well as in reading tasks, revealing persistent genetic effects on voice pitch across different contexts. Third, they are not dependent on mood: a heterogeneity test was not significant, suggesting consistent genetic effects between MDD patients and healthy people.
What does the finding of common genetic underpinnings in Mandarin Chinese (a language in which word meaning is conveyed by variation in pitch) and Icelandic (a language in which pitch does not play this role) reveal about the biology of speech and language? Marked differences in pitch patterns between tonal and non-tonal languages have been demonstrated in previous studies11,12, yet we found a cross-linguistic consistency in the influence of ABCC9 locus on pitch. We also observed a high degree of consistency in the direction of the genetic effects on pitch, with 98.99% of the SNPs exhibiting effects in the same direction at a P value threshold <\(1\times {10}^{-6}\). We think this consistency is because the analyzed phenotype, median F0, is primarily related to the non-linguistic production of speech, rather than to meaning. Spoken language, which involves cognition and emotional process, is more likely to be reflected in the variation of F0 over time during speech13, not to the mean or median values. Indeed, we found that it was the changes in pitch, not pitch itself, that were genetically correlated with MDD10.
Voice pitch is primarily modulated by fine changes in the tension of the vocal folds, which is mainly achieved by flexing the cricothyroid muscle, which causes the thyroid cartilage to tilt relative to the cricoid cartilage, thereby stretching the vocal folds11. Greater tension causes the vocal folds to vibrate at a higher frequency during voicing, producing a higher-pitch sound. The ABCC9 gene may influence voice pitch through hormonal pathways related to adrenal gland steroids, which produce several steroids known to influence voice pitch6,14. It can, alternatively, exert through nonhormonal mechanisms, affecting proteins in muscles related to the vocal fold or vocal tract6.
There are several limitations to our study. First, only women were included in the CONVERGE analysis. Although the Iceland study indicated that the effects of ABCC9 are irrespective of sex, it is unknown whether this finding is applicable to Chinese populations. Second, our analysis focused solely on mapping median F0. Other features, such as the variability of F0 and vowel acoustics, which represent different and possibly more crucial aspects of human vocal control ability, remain unexplored in our study.
In summary, through GWAS on Chinese women, we replicated the effects of a genetic locus at the ABCC9 gene on voice pitch. In combination with the Iceland study, we found two novel hits for pitch. These findings revealed common genetic effects on pitch across populations and languages.
Methods
Participants
The sample included 7654 women recruited from 55 provincial mental health centers in China as part of the CONVERGE (China, Oxford, and VCU Experimental Research on Genetic Epidemiology) study. Cases were aged between 30–60, with ≥ two episodes of MDD that met the DSM-IV criteria15. Control subjects, screened to exclude a history of MDD, were recruited from patients undergoing minor surgical procedures at general hospitals and individuals attending local community centers. Sample collection is described in detail in earlier work16,17,18. This study was approved by the Ethical Review Board of Oxford University (Oxford Tropical Research Ethics Committee) and local hospital review boards. All participants provided written informed consent.
Data collection
The voice data were obtained from semi-structured interviews, where the conversations between the subjects and the interviewer were recorded in hospital interview rooms. The interview was designed to assess psychiatric and demographic information in cases and controls. The length of case interviews was approximately three times longer than for controls because of a more detailed assessment of psychopathology and MDD risk factors (details of the interview protocol are provided in Supplementary Note 3). The recordings were not standardized and varied in quality and content. All recordings were listened to, and segments that contained only the patient’s voice at an adequate quality for analyses were identified. During this procedure, segments were manually labeled as to whether the speaker used a local dialect or had a non-standard accent for Mandarin Chinese, and a number was assigned to each recording to represent the level of background noise (four levels: 1. No noise; 2. Low noise; 3. Mild noise; 4. High noise). All segments from the same subject were concatenated into one, and downsampled to 8 kHz. Two postgraduate psychological students listened to all segments to ensure that no speech voice other than the interviewed subjects was included. All participants provided DNA samples for genetic analysis.
DNA was extracted from saliva samples using the Oragene protocol. Genotypes were acquired from low-coverage sequencing data from which SNPs were imputed. A sensitivity threshold of 90% to SNPs in the 1000 G Phase1 ASN panel was applied for SNP selection for imputation. Genotype likelihoods were calculated using a sample-specific binomial mixture model implemented in SNPtools (version 1.0)19, and imputation was performed using BEAGLE (version 3.3.2)20. A second round of imputation was performed with BEAGLE at biallelic SNPs polymorphic in the 1000 G Phase 1 ASN panel using the 1000 G Phase 1 ASN haplotypes as a reference panel. A final set of allele dosages and genotype probabilities was generated from these two datasets by replacing the results in the former with those in the latter at all sites imputed in the latter. We applied a conservative set of inclusion thresholds for SNPs for genome-wide association study: (a) p value for violation Hardy–Weinberg equilibrium p > 10-6, (b) Information score p > 0.9, (c) minor allele frequency >0.5%. Full details of the method and results are given in ref. 16.
Voice pitch calculation
We used the median F0 to measure pitch, which is more robust to outliers than mean values and is the same measure used in the Iceland study6. Given an audio segment, a time series of F0 values was computed using the Subharmonic Summation method and subsequently smoothed21. However, as our voice data were from spontaneous speech and contained more phonetic variation, we also calculated the first and the third quartiles of the F0 series, which were used for sensitivity tests. The calculation was implemented using the openSMILE package v2.4.222.
Heritability, genetic correlation, GWAS, and meta-analysis
The SNP-based heritability estimation used a GREML (generalized restricted maximum likelihood) method. GWAS was performed using a mixed model linear regression. Both were implemented in LDAK23. We adjusted for age, height, BMI, education level, marital status, occupation, social class, accent, noise level, total audio durations, the total number of audio segments concatenated, and 20 genetic principal components (PCs). The genetic correlation of pitch between cases and controls was calculated based on the individual genotype data, using the bivariate GREML method implemented in GCTA24.
Because of the expected differences in speech between the cases and controls10, we performed GWAS in the case and control subgroups separately. Then, we used meta-analyses to combine the two results. For cross-population analysis, we performed a meta-analysis of the summary statistics provided by the Iceland study, GWAS of our case subgroup, and GWAS of our control subgroup. The meta-GWAS were implemented in Metal25. Cochran’s Q-test26 was implemented for the heterogeneity test. The summary statistics of the Iceland study6 were lifted to GRCh37/hg19 and matched with our genotype data.
Polygenic score and binomial sign tests
We used the SNP associations from the Iceland study6 to construct PGS in the Chinese cohort. We first performed LD-based clumping (pairwise r2 > 0.5 in Chinese, 50 kb window) to remove markers from highly correlated SNP pairs. Then, we constructed PGS based on varying P value thresholds from \(5\times {10}^{-8}\) to 0.001 with an interval of \(5\times {10}^{-8}\). We assessed the predictive value of polygenic scores in the Chinese cohort by linear regression, with adjustment for the same covariates used in the GWAS analyses.
Using the same sets of SNPs and the P value thresholds ranging from \(5\times {10}^{-8}\) to 1 in the Iceland study, we applied a binomial sign test to determine whether the number of SNPs demonstrating consistent directions of allelic effects between Icelandic and Chinese was greater than expected by chance (that is, a one-sided test of whether this fraction is greater than 0.5).
Cross-ancestry fine-mapping
We used SuSiEx27,28 for fine-mapping analysis, which required two separate GWAS summary statistics in the single population and the corresponding LD matrixes. All variants in the ABCC9 locus that achieved genome-wide significance in the meta-analysis (Supplementary Data 1) were included. The LD matrixes were calculated using 1000 Genomes Project29 EAS/EUR samples as reference panel.
Statistics and reproducibility
We used the median F0 to measure pitch from 7654 Han Chinese Women. Saliva samples were collected and DNA was extracted using the Oragene protocol, as described above. GWAS on pitch was conducted in MDD cases and controls separately using a linear mixed model implemented in LDAK23, and then combined using a meta-analysis tool Metal25 (2011-03-25 version). The cross-population GWAS was conducted through a meta-analysis by combining the Chinese MDD cases, controls, and the Icelandic summary statistics, while the Icelandic results were lifted to GRCh37/hg19 and matched with the CONVERGE genotype data. Cross-population fine-mapping was implemented in SuSiEx27,28. The polygenic scores were calculated using a P value-based thresholding and clumping method implemented in PRSice30. In the one-sided binomial sign tests, we defined the null hypothesis as the probability of observing a consistent direction of genetic effects (as outlined in Table 1 for the total number of SNPs) being equal to 0.5. Conversely, the alternative hypothesis suggested that this probability is greater than 0.5.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The GWAS summary statistics of the median pitch in Chinese and the meta-GWAS are publicly available at FigShare (https://doi.org/10.6084/m9.figshare.24995963). The GWAS summary data for the median pitch in the Iceland study are available at Zenodo (https://doi.org/10.5281/zenodo.7152461). Due to the sensitive nature of the raw audio files and in adherence to privacy considerations, these files cannot be made publicly available. However, we are committed to facilitating scientific progress and transparency. Thus, secondary data derived from these audio files, specifically voice features, are available upon reasonable request. Researchers interested in accessing these data should contact the corresponding author, Jonathan Flint at JFlint@mednet.ucla.edu.
References
Simonyan, K. & Horwitz, B. Laryngeal motor cortex and control of speech in humans. Neuroscientist 17, 197–208 (2011).
Eising, E. et al. Genome-wide analyses of individual differences in quantitatively assessed reading- and language-related skills in up to 34,000 people. Proc. Natl Acad. Sci. USA 119, e2202764119 (2022).
Doust, C. et al. Discovery of 42 genome-wide significant loci associated with dyslexia. Nat. Genet. 54, 1621–1629 (2022).
den Hoed, J. & Fisher, S. E. Genetic pathways involved in human speech disorders. Curr. Opin. Genet. Dev. 65, 103–111 (2020).
Cavalcanti, J. C., Eriksson, A. & Barbosa, P. A. Multiparametric analysis of speaking fundamental frequency in genetically related speakers using different speech materials: some forensic implications. J. Voice https://doi.org/10.1016/j.jvoice.2021.08.013 (2021).
Gisladottir, R. S. et al. Sequence variants affecting voice pitch in humans. Sci. Adv. 9, eabq2969 (2023).
Yip, M. Tone (Cambridge Univ. Press, 2002).
Frick, R. W. Communicating emotion: the role of prosodic features. Psychol. Bull. 97, 412–429 (1985).
Sobin, C. Psychomotor symptoms of depression. Am. J. Psychiatry 15, 4–17 (1997).
Yazheng, D. et al. Unraveling the associations between voice pitch and major depressive disorder: a multisite genetic study. Preprint (Version 1) available at Research Square https://doi.org/10.21203/rs.3.rs-4135145/v1c (2024).
Dichter, B. K., Breshears, J. D., Leonard, M. K. & Chang, E. F. The control of vocal pitch in human laryngeal motor cortex. Cell 174, 21–31.e9 (2018).
Li, Y., Tang, C., Lu, J., Wu, J. & Chang, E. F. Human cortical encoding of pitch in tonal and non-tonal languages. Nat. Commun. 12, 1161 (2021).
Mozziconacci, S. Prosody and emotions. In Speech Prosody 2002, International Conference (2002).
Jost, L. et al. Associations of sex hormones and anthropometry with the speaking voice profile in the adult general population. J. Voice 32, 261–272 (2018).
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®) (American Psychiatric Association, 2013).
CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591 (2015).
Di, Y., Wang, J., Li, W. & Zhu, T. Using i-vectors from voice features to identify major depressive disorder. J. Affect. Disord. 288, 161–166 (2021).
Di, Y., Wang, J., Liu, X. & Zhu, T. Combining polygenic risk score and voice features to detect major depressive disorders. Front. Genet. 12, 2451 (2021).
Wang, Y., Lu, J., Yu, J., Gibbs, R. A. & Yu, F. An integrative variant analysis pipeline for accurate genotype/haplotype inference in population NGS data. Genome Res. 23, 833–842 (2013).
Browning, S. R. & Browning, B. L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007).
Eyben, F. Real-Time Speech and Music Classification by Large Audio Feature Space Extraction (Springer, 2015).
Eyben, F., Weninger, F., Gross, F. & Schuller, B. Recent developments in opensmile, the munich open-source multimedia feature extractor. In Proc. 21st ACM international Conference on Multimedia 835–838 (Association for Computing Machinery, 2013).
Speed, D., Holmes, J. & Balding, D. J. Evaluating and improving heritability models using summary statistics. Nat. Genet. 52, 458–462 (2020).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Cochran, W. G. The combination of estimates from different experiments. Biometrics 10, 101–129 (1954).
Yuan, K. et al. Fine-mapping across diverse ancestries drives the discovery of putative causal variants underlying human complex traits and diseases. Preprint at https://doi.org/10.1101/2023.01.07.23284293 (2023).
Wang, G., Sarkar, A., Carbonetto, P. & Stephens, M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Ser. B Stat. Methodol. 82, 1273–1300 (2020).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Euesden, J., Lewis, C. M. & O’Reilly, P. F. PRSice: polygenic risk score software. Bioinformatics 31, 1466–1468 (2015).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Acknowledgements
This work was funded by NIH grant MH-122596.
Author information
Authors and Affiliations
Contributions
J.F. conceived and organized this study. T.Z. and A.A. were responsible for overseeing the collection, cleaning, and analysis of the voice data. Y.D., J.W., V.R. and A.G. contributed to the voice data analysis. Y.D., J.M. and E.R. contributed to the genetic analysis. Y.D. and J.F. drafted the manuscript. All authors revised and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Veera Rajagopal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: George Inglis and Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Di, Y., Mefford, J., Rahmani, E. et al. Genetic association analysis of human median voice pitch identifies a common locus for tonal and non-tonal languages. Commun Biol 7, 540 (2024). https://doi.org/10.1038/s42003-024-06198-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06198-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.