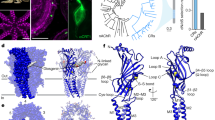
Abstract
The evolution of new traits enables expansion into new ecological and behavioural niches. Nonetheless, demonstrated connections between divergence in protein structure, function and lineage-specific behaviours remain rare. Here we show that both octopus and squid use cephalopod-specific chemotactile receptors (CRs) to sense their respective marine environments, but structural adaptations in these receptors support the sensation of specific molecules suited to distinct physiological roles. We find that squid express ancient CRs that more closely resemble related nicotinic acetylcholine receptors, whereas octopuses exhibit a more recent expansion in CRs consistent with their elaborated ‘taste by touch’ sensory system. Using a combination of genetic profiling, physiology and behavioural analyses, we identify the founding member of squid CRs that detects soluble bitter molecules that are relevant in ambush predation. We present the cryo-electron microscopy structure of a squid CR and compare this with octopus CRs1 and nicotinic receptors2. These analyses demonstrate an evolutionary transition from an ancestral aromatic ‘cage’ that coordinates soluble neurotransmitters or tastants to a more recent octopus CR hydrophobic binding pocket that traps insoluble molecules to mediate contact-dependent chemosensation. Thus, our study provides a foundation for understanding how adaptation of protein structure drives the diversification of organismal traits and behaviour.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Deep sequencing data are available via the Sequence Read Archive (SRA) repository under the BioProject accession code PRJNA906054. The CRB1 nucleotide sequence is available under NCBI accession OQ301567. Atomic model coordinates and cryo-EM density maps for the CRB1 structure have been deposited at the PDB under accession code 8EIZ and at the Electron Microscopy Data Bank under accession code EMD-28167, respectively.
References
van Giesen, L., Kilian, P. B., Allard, C. A. H. & Bellono, N. W. Molecular basis of chemotactile sensation in octopus. Cell 183, 594–604.e14 (2020).
Noviello, C. M. et al. Structure and gating mechanism of the α7 nicotinic acetylcholine receptor. Cell 184, 2121–2134 (2021).
Albertin, C. B. et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224 (2015).
Mollo, E., Garson, M. J., Polese, G., Amodeo, P. & Ghiselin, M. T. Taste and smell in aquatic and terrestrial environments. Nat. Prod. Rep. 34, 496–513 (2017).
Mollo, E. et al. Sensing marine biomolecules: smell, taste, and the evolutionary transition from aquatic to terrestrial life. Front. Chem. 2, 92 (2014).
Hanlon, R. T. & Messenger, J. B. Cephalopod Behaviour (Univ. Cambridge, 1996).
Graziadei, P. Receptors in the sucker of the cuttlefish. Nature 203, 384–387 (1964).
Bargmann, C. I. Comparative chemosensation from receptors to ecology. Nature 444, 295–301 (2006).
Basak, S., Gicheru, Y., Rao, S., Sansom, M. S. P. & Chakrapani, S. Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3A receptor. Nature 563, 270–274 (2018).
Dwyer, T. M., Adams, D. J. & Hille, B. The permeability of the endplate channel to organic cations in frog muscle. J. Gen. Physiol. 75, 469–492 (1980).
Polovinkin, L. et al. Conformational transitions of the serotonin 5-HT3 receptor. Nature 563, 275–279 (2018).
Zhang, Y. et al. Asymmetric opening of the homopentameric 5-HT3A serotonin receptor in lipid bilayers. Nat. Commun. 12, 1074 (2021).
Brejc, K. et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature 411, 269–276 (2001).
Jiao, Y., Cao, Y., Zheng, Z., Liu, M. & Guo, X. Massive expansion and diversity of nicotinic acetylcholine receptors in lophotrochozoans. BMC Genom. 20, 937 (2019).
Albertin, C. B. et al. Genome and transcriptome mechanisms driving cephalopod evolution. Nat. Commun. 13, 2427 (2022).
Zhang, Y. et al. The genome of Nautilus pompilius illuminates eye evolution and biomineralization. Nat. Ecol. Evol. 5, 927–938 (2021).
Brams, M. et al. Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT3 receptor via a common vestibule site. eLife 9, e51511 (2020).
Morales-Perez, C. L., Noviello, C. M. & Hibbs, R. E. Manipulation of subunit stoichiometry in heteromeric membrane proteins. Structure 24, 797–805 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D 74, 814–840 (2018).
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
The PyMOL Molecular Graphics System v.2.4.1 (Schrodinger, 2015).
UniProt, C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Pei, J., Kim, B. H. & Grishin, N. V. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Klesse, G., Rao, S., Sansom, M. S. P. & Tucker, S. J. CHAP: a versatile tool for the structural and functional annotation of ion channel pores. J. Mol. Biol. 431, 3353–3365 (2019).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Tian, W., Chen, C., Lei, X., Zhao, J. & Liang, J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 46, W363–W367 (2018).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Eddy, S. R. Profile hidden Markov models. Bioinformatics 14, 755–763 (1998).
Madden, T. in The NCBI Handbook 2nd edn (eds McEntyre, J. & Ostell, J.) Ch. 16 (2002).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Dos Reis, M. & Yang, Z. Bayesian molecular clock dating using genome-scale datasets. Methods Mol. Biol. 1910, 309–330 (2019).
Tanner, A. R. et al. Molecular clocks indicate turnover and diversification of modern coleoid cephalopods during the Mesozoic Marine Revolution. Proc. R. Soc. B 284, 20162818 (2017).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T.-Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Revell, L. J. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012).
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006).
Tinevez, J.-Y. et al. TrackMate: An open and extensible platform for single-particle tracking. Methods 15, 80–90 (2017).
Jurrus, E. et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Acknowledgements
We thank C. Winkler for providing octopuses; B. Grasse of the Cephalopod Program and the Marine Resource Center at the MBL for squid; B. Walsh for animal husbandry; C. Heacock for help with nerve recordings; R. Lines for assistance with behavioural experiments; A. Grearson for photographs and illustrations; S. Burke for contributions to early stages of the project; J. Teng for preliminary functional tests; R. Cabuco and L. Baxter for baculovirus production; and C. Noviello for screening grids. Single-particle cryo-EM grids were screened at the University of Texas Southwestern Medical Center Cryo-Electron Microscopy Facility, which is supported by the CPRIT Core Facility Support Award RP170644. A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. This research was further supported by grants to N.W.B. from the New York Stem Cell Foundation, Searle Scholars Program and the NIH (R35GM142697 and R01NS129060); NIH grants to R.E.H. (R01NS120496 and R01NS129060) and to G.K. (F32DA047848); and support from the American Heart Association to J.J.K. (20POST35200127) and from the National Science Foundation to C.A.H.A. (NSF-PRFB 2010728).
Author information
Authors and Affiliations
Contributions
C.A.H.A., W.A.V.-M., L.v.G., P.B.K. and N.W.B. contributed to molecular, cellular and organismal studies. G.K., J.J.K., X.B. and R.E.H. contributed to biochemical and structural analyses. All of the authors were involved with writing or reviewing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Hugues Nury, Oleg Simakov and Harold Zakon for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Comparative chemotactile behaviour.
a, Squid have two long tentacles for catching prey, in addition to eight arms like octopus. b, Octopuses explore their home tank, moving throughout recordings while squid are stationary and often buried. n = 8-9 trials. c, Octopuses move toward their prey for capture while squid wait for prey to approach and then strike. n = 4 trials, p < 0.0001 for octopus versus squid movement, two-tailed Student’s t-test. d, Octopuses spread their arms in relatively dark conditions and contract in the light. n = 10 trials, p < 0.0001 for octopus versus squid movement, two-tailed Student’s t-test. e, Octopuses pounce on and consume live crabs from a distance in the light and must first touch freshly dead crabs before consuming. In the dark, octopuses must first touch live or dead crabs before pouncing and consuming. These results suggest octopuses use somatosensation for predation in the absence of strong visual cues. n = 9–13 trials. f, Squid use ambush predation to hide and strike shrimp in light or relatively dark conditions with a similar success rate but require that prey are at a much closer distance in the dark. These results suggest squid primarily use vision to strike prey. p < 0.0001 for strike distance in light versus dark, two-tailed Student’s t-test. n = 18–24 trials. Data represented as mean ± SEM.
Extended Data Fig. 2 Comparative chemosensory cell electrical properties.
a, Quantification of chemosensory cells from octopus or squid that responded to 3 kDa fish and shrimp extract in voltage- or current-clamp. b, Shrimp-sensitive octopus chemoreceptor cell currents were blocked by 1 mM mecamylamine. p < 0.05, two-tailed Student’s t-test. n = 7 cells. Scale bar: 5 μm. c, Representative fish-evoked action potentials in an octopus chemosensory cell. d, 1 μM tetrodotoxin (TTX) blocked voltage-gated inward currents (INav) in octopus chemosensory cells. n = 6-11 cells, p < 0.0001, two-way ANOVA with post hoc Bonferroni test. e, Octopus chemosensory cell INav voltage dependence: Va1/2 = −5.1 ± 0.7 mV, Vi1/2 = −38.0 ± 1.8 mV, n = 4. f, Octopus chemosensory cell INav activation and inactivation kinetics. n = 6. g, 1 μM tetrodotoxin (TTX) blocked voltage-gated inward currents (INav) in squid chemosensory cells. n = 6–11 cells, p < 0.0001, two-way ANOVA with post hoc Bonferroni test. h, Squid chemosensory cell INav voltage dependence: Va1/2 = −6.5 ± 0.8 mV, Vi1/2 = −40.3 ± 1.3 mV, n = 4 cells. i, Squid chemosensory cell INav activation and inactivation kinetics. n = 6 cells. j, Voltage-gated outward currents measured in the presence of intracellular potassium (IKV) in squid and octopus chemosensory cells. n = 5 cells for squid and 6 for octopus. k, Both squid and octopus chemosensory cells produced action potentials in response to current injection. Squid had slightly higher frequency in response to the same injection step. n = 5 cells for squid and 7 for octopus. l, comparison of spike amplitude and input resistance in squid and octopus chemosensory cells. n = 5 cells for squid and 7 for octopus. Data represented as mean ± SEM.
Extended Data Fig. 3 Squid CR properties.
a, Alignment of predicted loops A, B and C from analysed octopus and cuttlefish CRs and the human α7 nicotinic acetylcholine receptor demonstrated that CRs lack most residues that contribute to the canonical acetylcholine (ACh) binding side (highlighted in red). b, Squid CRs localized to the sensory epithelium of arm suckers (left, merge in Fig. 1) and tentacle suckers (right), as visualized by RNAscope in situ hybridization. Nuclei were stained with DAPI (blue). Representative of 3 animals. c, Expressed octopus (CR518, CRT1) and cuttlefish CRs (CR192, CRB1) were insensitive to acetylcholine (ACh, 1 mM) but robustly responded to fish or shrimp extract. Extract responses were blocked by mecamylamine (1 mM) and were not observed in untransfected cells. n = 6–7 cells, p < 0.0001 for extract responses versus mecamylamine, two-way ANOVA with post-hoc Tukey test. d, Octopus CRT1 exhibited dose-dependent sensitivity to the terpene costunolide, while squid CRB1 was insensitive. Octopus CRT1 EC50 = 30.8 μM, 95% CI = 30.0 – 33.9 μM, n = 7 cells for octopus and 5 for squid. Data represented as mean ± SEM. e, Minimal desensitization was measured in response to low concentration of denatonium while higher concentrations produced inhibition with large wash-off currents that were absent at positive voltage. These properties are consistent with moderate pore block. p < 0.0001 for concentration, two-tailed student’s t-test (n = 6). Data represented as mean ± SEM.
Extended Data Fig. 4 Comparative squid and octopus nerve and arm chemosensory responses.
a, (Top) arm and tentacle axial nerves stained with anti-horseradish peroxidase antibody (purple) and nuclear stain (DAPI, blue). Representative of at least 3 animals. Scale bars: 1 mm. (Bottom) Corresponding nerve recordings in response to the indicated stimuli: control (sea water), 3 kDa fish extract, 1 mM nootkatone, 1 mM denatonium. b, Amputated arm behaviour in response to control (sea water), 3 kDa fish extract, 1 mM denatonium, or 1mM nootkatone. c, Heat map of normalized axial nerve and arm responses. Octopus and squid were sensitive to terpenes but only squid arms and tentacles responded to denatonium. p < 0.05 for denatonium sensitivity in squid, n = 3 arms. d, Representative axial nerve recordings from longfin (D. pealii) and bobtail squid (E. berryi) arms in response to control (sea water), 3 kDa fish extract, 1 mM denatonium. e, Heat map of normalized axial nerve responses in longfin and bobtail squid. f, Normalized surface area of octopus arms touching agar coated floors was reduced on the side containing polygodial (100 μM, n = 7). Octopuses touched both control sides the same. p < 0.01, paired Student’s t test, n = 8 trials per condition. g, Squid touched agar-coated floors with the same frequency and surface area when both sides were coated with sea water-filled agar or one side contained denatonium (also see Fig. 2). n = 7 trials.
Extended Data Fig. 5 EM data processing.
a-d, 2D slices through TMDs from reconstructions of CRB1 in different membrane mimetics. e, Representative cryo-electron micrograph of CRB1 in GDN detergent micelles from a dataset of 16,042 dose-fractionated micrographs. Scale bar = 100 nm. f, Projection images from the selected 2D classes. g, 3D classification results; good 3D classes selected for separate processing are boxed in red and yellow. h-i, 3D classification results; 3D classes selected are boxed in red, and particles from selected 3D classes were aligned and combined for 3D refinement. j, 3D classification results using local angular searches; best 3D class with strong TMD density boxed in red was selected for additional 3D classification. k, TMD-focused 3D classification results; particles from best 3D class (boxed in red) were used in final refinement. l, 3D reconstructed map from the final 3D classification, which is shown in both side and top views. Density of M1-M4 for each subunit is outlined with a black dashed line. m, Sharpened map coloured by local resolution. n, Half-map FSC plot for masked and unmasked maps with global resolution indicated at FSC = 0.143.
Extended Data Fig. 6 Representative density map segmentation.
a, Cryo-EM density map of CRB1 for representative adjacent subunits coloured in gold and yellow. Density map of denatonium coloured in cyan and waters in red at a threshold level of 0.03. b, Orthosteric binding site of CRB1 boxed in a, where residues within 5 Å of denatonium and negatively charged residues near waters are shown as sticks. c, Calculated interface areas and interaction energies (ΔiG) for protein and denatonium using PDBePISA38. Calculated solvent accessible area and volume of the binding pocket for CRB1 using CASTp3.039. d–i, Cryo-EM density segments of Loops A-F at the orthosteric binding site at a threshold level of 0.03. j–o, Cryo-EM density segments of Cys-loop, β8-β9-loop, β1-β2-loop, β4-β5-loop, M1M2 loop, and M2M3 loop at a threshold level of 0.03. p–t, Cryo-EM density segments of M1-M3 and two M4 helices (chains C and E) at a threshold level of 0.03.
Extended Data Fig. 7 Squid CR ion permeation pathway.
a, Pore radii as a function of distance along the pore axis; CRB1 is coloured in yellow and CRT1 in blue. Structures were aligned using the M2 helix Glu-1ʹ at the bottom of M2, which is defined as y = 0. b, CRB1 ion permeation pathway coloured by hydrophobicity37 with D102 and F46 indicated as spheres, the two most constricted points in the ECD; front subunit removed for clarity. c, Top view of CRB1 shows negatively charged D102 points toward the channel axis. d, Representative current-voltage (I-V) relationships of WT and D102A mutant CRB1 channels in response to denatonium during equimolar cation substitution. e, D102A did not change permeation but did affect current amplitude measured at +60 mV in the presence of all permeant cations. Outward currents in external NMDG+ were the same in WT versus D102A. n = 7–10 cells, p < 0.0001 two-way ANOVA with post-hoc Bonferroni test. Data represented as mean ± SEM. f, Structure-based sequence alignment of β4-β5 loop (Ω loop) for CRB1, CRT1, and 5-HT3A (PDB: 6NP0). g, Comparison of Ω loop conformation of CRB1 coloured in pink and 5-HT3A (Ω-open) coloured in grey; D102 of CRB1 is shown as sticks. h, Comparison of Ω loop conformation of CRT1 coloured in pink and 5-HT3A (Ω-open) coloured in grey; E104 of CRT1 is shown as sticks. i, Structure-based sequence alignment of Ω loop for α7 (PDB: 7KOX), α3 (PDB: 6PV7), α4 nicotinic receptor (PDB: 5KXI). j, Comparison of Ω loop conformation of α7 nicotinic receptor coloured in light blue and 5-HT3A (Ω-open) coloured in grey; E97 of α7 is shown as sticks. k, Comparison of Ω loop conformation of α7 nicotinic receptor coloured in light blue and α4 nicotinic receptor (Ω-in) coloured in light green. m,n, Comparison of pore shapes analysed by HOLE236. o,p, Comparison of relative symmetry in the TMD (o) and ECD (p) for CRT1 and CRB1. q,r, α7 receptor electrostatics analysed by APBS56, where r is a cutaway to show electrostatics of the permeation pathway, calculated with 140 mM NaCl. s–v, Electrostatics for CRB1 and CRT1, calculated with 0.5 M NaCl.
Extended Data Fig. 8 Superposition of subunits from CRs and α7 nicotinic receptor.
a–e, Superposition of subunits within CRB1, aligned by ECD region using UCSF Chimera. The r.m.s.d values in Å are for Cα atoms over the whole chain, calculated by secondary structure-based alignment using Coot. Chain IDs are indicated in parentheses. f–j, Subunits of CRB1 are compared to a subunit of CRT1. k–o, Subunits of CRB1 are compared to a subunit of α7 nicotinic receptor (activated state, PDB:7KOX).
Extended Data Fig. 9 Squid CR hydrophobic cluster and hydrogen bond network.
a, Single subunit structure of squid CR predicted by AlphaFold57 and coloured by confidence. b, Experimental cryo-EM structure of single subunit with two disulfide bonds shown as spheres. c, Details of CRB1 boxed in b, and a 180° rotated view. Hydrophobic residues contacting the two disulfide bonds are shown as sticks. d, Structure-based sequence alignment of β8-β9 loop and Loop C of CRB1, CRT1, and human α7 nicotinic acetylcholine receptor. A novel disulfide bond in CRB1 is highlighted in a yellow box, and the corresponding residue on the β8-β9 loop in CRT1 and α7 is indicated in a magenta-coloured box. e, Hydrogen bond network between β9, β10, β7 and β6 in CRB1 shown as black dashed line. f, Hydrogen bond network between β9, β10, and β7 in CRT1 at the same view as e. The β9 strand shifts upward as indicated by the magenta arrow due to insertion of the Y166 sidechain between β9 and β10, which breaks the hydrogen bond network in this region. g, Hydrogen bond network between β9, β10, and β7 in α7 in the same orientation as e. h-m, In the TMD, density for the most peripheral M4 helix was notably absent in CRT1, in all tested detergents, but it was predicted to be present by AlphaFold. h, j, and l show predicted structures of TMD bundles for CRB1, α7, and CRT1. i, k, and m show interactions as lines from PDBePISA server analysis; solid lines indicate interactions found in both the experimental structures and in the AlphaFold model; dashed lines indicate those found in only the experimental structures. In panel m, solid lines indicate interactions found only in the AlphaFold model. Analysis of molecular interactions among M1/M3/M4 helices from the experimental and predicted models sheds light on the basis for the presence or absence of an ordered M4 helix. In CRB1 and α7, plentiful aromatic contacts among helices stabilize M4 packing in the TMD bundle, while the most recently diverged CRT1 lacks the abundance of hydrophobic contacts.
Extended Data Fig. 10 Comparative squid and octopus CR phylogeny.
a, Phylogenetic tree of acetylcholine-receptor-like protein sequences from Octopus bimaculoides (Obim), Octopus sinensis (Osin), Sepioloidea lineolata (Slin), Doryteuthis pealeii (Dpea), Euprymna berryi (Eber), Nautilus pompilius (Npom), and Lottia gigantea (Lgig) showing that CRs diverged from acetylcholine-like receptors, are unique to coleoid cephalopods, and comprise three major lineages: CRB (CR-Bitter), CRT (CR-Terpenes), and CRX (CR with unknown ligands). b, Density distributions of neutral genetic diversity (fourfold degenerate site distances, 4DTv) estimated for all pairs of genes between CRB and CRT clades. Dotted lines correspond to the mode of distributions for pairwise comparisons between CRTs and CRBs, suggesting CRTs have undergone a more recent diversification. c, Densitree of the distribution of bootstrap topologies for the CR clade showing that CRB is robustly supported as the earliest divergent lineage of CRs and sister to the CRT-CRX clade.
Supplementary information
Supplementary Data
Sequence alignment of CRT1, CRB1 and other Cys-loop receptor subunits.
Supplementary Table 1
Tissue-specific de novo transcriptomes for S. lineolata, D. pealeii and E. berryi.
Supplementary Video 1
Octopus prey capture. Octopuses moved around their tank to find and capture crabs. Video in real time
Supplementary Video 2
Squid prey capture. Squids buried themselves and strike when shrimps moved close enough. 0.5× speed.
Supplementary Video 3
Octopus chemotactile behaviour. Octopuses reduced contact with 100 μM polygodial-infused 1.5% agarose versus sea water control. 4× speed.
Supplementary Video 4
Squid chemotactile behaviour. Squid behaved similarly when their body contacted 1 mM denatonium-infused 1.5% agarose or sea water control. 4× speed.
Supplementary Video 5
Agonist-binding site of CRB1. Animation showing CRB1 bound to denatonium. Magnified view shows orthosteric binding pocket with ligand-coordinating residues highlighted.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kang, G., Allard, C.A.H., Valencia-Montoya, W.A. et al. Sensory specializations drive octopus and squid behaviour. Nature 616, 378–383 (2023). https://doi.org/10.1038/s41586-023-05808-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-05808-z
This article is cited by
-
Welcome to the nursery
Nature Methods (2023)
-
Structural basis of sensory receptor evolution in octopus
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.