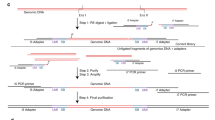
Abstract
The discovery of somatic mutations in cancer tissue is extremely laborious, time-consuming and costly. In an evaluation comparing mismatch repair detection (MRD) against Sanger sequencing for somatic-mutation detection, we found that MRD had a specificity of 96% and a sensitivity of 92%. Our results showed that MRD is a robust and cost-effective alternative to Sanger sequencing for identifying somatic mutations in human tumors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Davies, H. et al. Cancer Res. 65, 7591–7595 (2005).
Sjoblom, T. et al. Science 314, 268–274 (2006).
Greenman, C. et al. Nature 446, 153–158 (2007).
Faham, M. & Cox, D.R. Genome Res. 5, 474–482 (1995).
Faham, M. et al. Proc. Natl. Acad. Sci. USA 102, 14717–14722 (2005).
Nelson, M.R. et al. Genome Res. 14, 1664–1668 (2004).
Fakhrai-Rad, H. et al. Genome Res. 14, 1404–1412 (2004).
Gresham, D. et al. Science 311, 1932–1936 (2006).
Forbes, S. et al. Br. J. Cancer 94, 318–322 (2006).
Orlicky, S. et al. Cell 112, 243–256 (2003).
Rajagopalan, H. et al. Nature 428, 77–81 (2004).
Lee, J.W. et al. Eur. J. Cancer 42, 2369–2373 (2006).
Kwak, E.L. et al. Gynecol. Oncol. 98, 124–128 (2005).
Calhoun, E.S. et al. Am. J. Pathol. 163, 1255–1260 (2003).
Acknowledgements
We thank J. Peña for graphical help with Figure 1, the DNA sequencing lab for thoughtful discussions, D. Kenski and M. Eby for careful review of the manuscript and G. Cavet, J. Kaminker, S. Guerrero, J. Yuan and S. Lohr for help with informatics analysis and database management.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
All authors are employed by either Genentech or Affymetrix, and own stock in their respective companies.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1, 2, 4–6, Supplementary Methods. (PDF 608 kb)
Supplementary Table 3
Amplicon call rates by sample. (XLS 846 kb)
Rights and permissions
About this article
Cite this article
Peters, B., Kan, Z., Sebisanovic, D. et al. Highly efficient somatic-mutation identification using Escherichia coli mismatch-repair detection. Nat Methods 4, 713–715 (2007). https://doi.org/10.1038/nmeth1081
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1081