Featured
-
-
Letter |
 The heat released during catalytic turnover enhances the diffusion of an enzyme
The heat released during catalytic turnover enhances the diffusion of an enzyme
It has been traditionally assumed that the heat released during a single enzymatic catalytic event does not perturb the enzyme in any way; however, here single-molecule fluorescence correlation spectroscopy is used to show that, for enzymes that catalyse chemical reactions with large reaction enthalpies, the heat released at the protein's active site during catalysis transiently displaces the protein's centre-of-mass, essentially giving rise to a recoil effect that propels the enzyme.
- Clement Riedel
- , Ronen Gabizon
- & Carlos Bustamante
-
Letter |
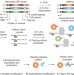 Multiplex single-molecule interaction profiling of DNA-barcoded proteins
Multiplex single-molecule interaction profiling of DNA-barcoded proteins
Single-molecular-interaction-sequencing involves attaching DNA barcodes to proteins, assaying these barcoded proteins en masse in an aqueous solution, followed by immobilization in a polyacrylamide film and amplifying and analysing the barcoding DNAs—the method allows for precise protein quantification and simultaneous interrogation of molecular binding affinity and specificity.
- Liangcai Gu
- , Chao Li
- & George M. Church
-
Letter |
 Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA
Histone H4 tail mediates allosteric regulation of nucleosome remodelling by linker DNA
A nucleosome-spacing mechanism for human ATP-dependent chromatin assembly and remodelling factor (ACF).
- William L. Hwang
- , Sebastian Deindl
- & Xiaowei Zhuang
-
Letter |
 Dynamic pathways of −1 translational frameshifting
Dynamic pathways of −1 translational frameshifting
To investigate the mechanism of frameshifting during messenger RNA translation, a technique was developed to monitor translation of single molecules in real time using Förster resonance energy transfer (FRET); ribosomes were revealed to pause tenfold longer than usual during elongation at the frameshifting sites.
- Jin Chen
- , Alexey Petrov
- & Joseph D. Puglisi
-
Article |
 Protein-guided RNA dynamics during early ribosome assembly
Protein-guided RNA dynamics during early ribosome assembly
Three-colour fluorescence resonance energy transfer and molecular dynamics simulations are used to study the events occurring early in assembly of the 30S ribosome; within a non-native intermediate S4 ribosomal protein–16S RNA structure, S4 is capable of altering the RNA helix dynamics to facilitate conformation changes that enable subsequent protein binding.
- Hajin Kim
- , Sanjaya C. Abeysirigunawarden
- & Sarah A. Woodson
-
Article |
 DNA interrogation by the CRISPR RNA-guided endonuclease Cas9
DNA interrogation by the CRISPR RNA-guided endonuclease Cas9
This study defines how a short DNA sequence, known as the PAM, is critical for target DNA interrogation by the CRISPR-associated enzyme Cas9 — DNA melting and heteroduplex formation initiate near the PAM and extend directionally through the remaining target sequence, and the PAM is also required to activate the catalytic activity of Cas9.
- Samuel H. Sternberg
- , Sy Redding
- & Jennifer A. Doudna
-
Letter |
 Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit
Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit
A sub-nanometre reconstruction of a 40S complex containing eIF3 and a hepatitis C virus (HCV)-like internal ribosome entry site (IRES) shows that the IRES displaces eIF3 from the 40S and sequesters it to gain access to the 40S subunit.
- Yaser Hashem
- , Amedee des Georges
- & Joachim Frank
-
Letter |
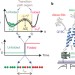 Single-molecule fluorescence probes dynamics of barrier crossing
Single-molecule fluorescence probes dynamics of barrier crossing
Here the Kramers diffusion coefficient and free-energy barrier are characterized for the first time through single-molecule fluorescence measurements of the temperature- and viscosity-dependence of the transition path time for protein folding.
- Hoi Sung Chung
- & William A. Eaton
-
Letter |
 Unsynchronised subunit motion in single trimeric sodium-coupled aspartate transporters
Unsynchronised subunit motion in single trimeric sodium-coupled aspartate transporters
Glutamate transporters are integral membrane proteins that facilitate neurotransmitter uptake from the synaptic cleft into the cytoplasm of glial cells and neurons, the mechanism of transport involves transitions between extracellular- and intracellular-facing conformations; here the authors used single-molecule fluorescence resonance energy transfer imaging to directly observe conformational dynamics in trimers of a bacterial homologue of glutamate transporters that was embedded in the membrane.
- Guus B. Erkens
- , Inga Hänelt
- & Antoine M. van Oijen
-
Letter |
 DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate
DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate
The bacterial RecBCD helicase/nuclease shows broad, and apparently static, heterogeneity in the unwinding rate manifest by individual molecules: here it is shown that transiently halting an enzyme during processive translocation allows for a change, most likely conformational, such that the velocity of the molecule after pausing can fall anywhere within the spectrum of rates seen for a population.
- Bian Liu
- , Ronald J. Baskin
- & Stephen C. Kowalczykowski
-
Letter |
 Reshaping of the conformational search of a protein by the chaperone trigger factor
Reshaping of the conformational search of a protein by the chaperone trigger factor
The bacterial chaperone named trigger factor is found to stabilize protein folding intermediates that eventually convert to the native state, suggesting that chaperones play a direct role in instructing protein folding.
- Alireza Mashaghi
- , Günter Kramer
- & Sander J. Tans
-
Letter |
 Transport dynamics in a glutamate transporter homologue
Transport dynamics in a glutamate transporter homologue
Single-molecule fluorescence resonance energy transfer imaging of a bacterial glutamate transporter reveals how the transport domains move.
- Nurunisa Akyuz
- , Roger B. Altman
- & Olga Boudker
-
Letter |
 Modulation of allostery by protein intrinsic disorder
Modulation of allostery by protein intrinsic disorder
Single-molecule FRET is used to examine how an intrinsically disordered protein, the adenovirus E1A oncoprotein, interacts with two different protein partners (the pocket domain of pRb and the TAZ2 domain of CBP/p300); the biophysical behaviour of E1A depends on whether the N-terminal region and/or the CR2 region of E1A is free to interact with potential protein partners or whether they are ‘masked’ (that is, via their absence or a pre-existing interaction with another protein partner).
- Allan Chris M. Ferreon
- , Josephine C. Ferreon
- & Ashok A. Deniz
-
News & Views |
 All clear for ribosome landing
All clear for ribosome landing
The discovery of a dramatic structural rearrangement that is stabilized by an RNA scaffold helps to explain how nascent proteins are delivered for export from the cell cytoplasm. See Letter p.271
- Harris D. Bernstein
-
Letter |
 Activated GTPase movement on an RNA scaffold drives co-translational protein targeting
Activated GTPase movement on an RNA scaffold drives co-translational protein targeting
Single-molecule fluorescence microscopy techniques are used to elucidate features of the highly conserved protein-targeting machinery known as the signal recognition particle (SRP); the long SRP RNA is shown to be crucial for correct timing and precision of cargo handover to the protein-translocation machinery, a finding that could help to explain how other ribonucleosome complexes function during complex cellular processes.
- Kuang Shen
- , Sinan Arslan
- & Shu-ou Shan
-
News & Views |
 Molecular hurdles cleared with ease
Molecular hurdles cleared with ease
Single-molecule studies reveal that a ring-like enzyme that encircles and 'slides' along one strand of duplex DNA, separating it from the other strand, overcomes molecular barriers in its path by transiently opening its ring. See Article p.205
- Michael A. Trakselis
- & Brian W. Graham
-
Article |
 Bypass of a protein barrier by a replicative DNA helicase
Bypass of a protein barrier by a replicative DNA helicase
Single-molecule and ensemble assays are used to show that large T antigen, the replicative DNA helicase of the simian virus 40 (SV40), unwinds DNA as a single hexamer by steric exclusion and is able to bypass covalent DNA–protein crosslinks.
- Hasan Yardimci
- , Xindan Wang
- & Johannes C. Walter
-
News & Views |
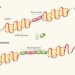 A glimpse of molecular competition
A glimpse of molecular competition
Single-molecule studies reveal how the DNA-repair protein RecA overcomes competition from another protein to bind to single-stranded DNA, and how other mediator proteins assist in this process. See Letter p.274
- Susan T. Lovett
-
Letter |
 Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA
Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA
Single-molecule analysis of RecA filament assembly on its in vivo substrate, SSB-coated single-stranded DNA, reveals that a dimer of RecA is required for nucleation, and is followed by bidirectional growth of the filament through monomer addition; the recombination mediator RecOR accelerates nucleation and growth, and the addition of RecF further stimulates nucleation.
- Jason C. Bell
- , Jody L. Plank
- & Stephen C. Kowalczykowski
-
Letter |
 Heterogeneous pathways and timing of factor departure during translation initiation
Heterogeneous pathways and timing of factor departure during translation initiation
A single-molecule approach is used to investigate the kinetics of assembly of the translation initiation complex, revealing that there is more than one pathway by which the necessary factors can assemble.
- Albert Tsai
- , Alexey Petrov
- & Joseph D. Puglisi
-
Letter |
 Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search
Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search
The search for DNA homology is vital to recombinational DNA repair and occurs by intersegment contact sampling wherein the three-dimensional conformational state of the double-stranded DNA target and the length of the homologous RecA–single-stranded DNA filament have important roles.
- Anthony L. Forget
- & Stephen C. Kowalczykowski
-
Letter |
 ATP-induced helicase slippage reveals highly coordinated subunits
ATP-induced helicase slippage reveals highly coordinated subunits
- Bo Sun
- , Daniel S. Johnson
- & Michelle D. Wang
-
Review Article |
 Central dogma at the single-molecule level in living cells
Central dogma at the single-molecule level in living cells
- Gene-Wei Li
- & X. Sunney Xie
-
Review Article |
 Nuclear export dynamics of RNA–protein complexes
Nuclear export dynamics of RNA–protein complexes
- David Grünwald
- , Robert H. Singer
- & Michael Rout
-
Letter |
 Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins
Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins
- Madeleine B. Borgia
- , Alessandro Borgia
- & Jane Clarke
-
Article |
 Probing cellular protein complexes using single-molecule pull-down
Probing cellular protein complexes using single-molecule pull-down
- Ankur Jain
- , Ruijie Liu
- & Taekjip Ha
-
Letter |
 Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue
Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue
- Yongfang Zhao
- , Daniel S. Terry
- & Jonathan A. Javitch
-
Article |
 Single-molecule analysis of Mss116-mediated group II intron folding
Single-molecule analysis of Mss116-mediated group II intron folding
DEAD-box helicases use ATP hydrolysis to unwind duplex RNA and facilitate RNA or RNA–protein remodelling. One such helicase is Mss116, which targets a particular group II intron in RNA. Here, single-molecule fluorescence was used to monitor the effect of Mss16 on a minimal construct containing this intron. The data show that Mss16 stimulates the sampling of different folded states of the RNA. Moreover, the helicase promotes RNA folding through discrete ATP-independent and ATP-dependent steps.
- Krishanthi S. Karunatilaka
- , Amanda Solem
- & David Rueda
-
Letter |
 Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking
Selectivity mechanism of the nuclear pore complex characterized by single cargo tracking
Nuclear pore complexes selectively transport cargos across the nuclear envelope. Here, a nuclear transport assay has been developed that allows the movement of single cargo proteins to be followed in real time. A succession of transport substeps is observed, and the NPC is found to be functionally asymmetric to importing cargos. The study provides insight into the mechanism of selective transport through the NPC.
- Alan R. Lowe
- , Jake J. Siegel
- & Jan T. Liphardt
-
Letter |
 A mechanically stabilized receptor–ligand flex-bond important in the vasculature
A mechanically stabilized receptor–ligand flex-bond important in the vasculature
Here, a new type of behaviour of receptor–ligand bonds has been identified, by using a new method that links receptor and ligand in a single molecule to measure binding and unbinding. The binding of von Willebrand factor to the glycoprotein Ib α subunit on the surface of platelets is important for coagulation. This receptor–ligand bond is now shown to have two distinct states, one seen at low force and a second that has greater force resistance. This has implications for how increased blood flow activates platelet plug formation.
- Jongseong Kim
- , Cheng-Zhong Zhang
- & Timothy A. Springer
-
Research Highlights |
Biophysics: Molecular carnival ride
-
News & Views |
 Transporter in the spotlight
Transporter in the spotlight
Membrane transporter proteins switch between conformational states to move substrates across membranes. The transition between these states can now be studied using single-molecule experiments.
- Nathan K. Karpowich
- & Da-Neng Wang

 Transport domain unlocking sets the uptake rate of an aspartate transporter
Transport domain unlocking sets the uptake rate of an aspartate transporter