Featured
-
-
Letter |
 Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex
Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex
The structure of the METTL3–METTL14 complex, which mediates N6-adenosine methylation of RNA, suggests that the METTL3 subunit is the catalytic core while METTL14 serves to bind RNA.
- Xiang Wang
- , Jing Feng
- & Ping Yin
-
Letter |
 The crystal structure of Cpf1 in complex with CRISPR RNA
The crystal structure of Cpf1 in complex with CRISPR RNA
The crystal structure of monomeric Lachnospiraceae bacterium Cpf1 protein bound to CRISPR RNA is presented, establishing a framework for engineering LbCpf1 to improve its efficiency and specificity for genome editing.
- De Dong
- , Kuan Ren
- & Zhiwei Huang
-
Letter |
 The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA
The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA
The CRISPR-associated protein Cpf1 from Francisella novicida is a novel enzyme with specific, dual-endoribonuclease–endonuclease activities in precursor crRNA processing and crRNA-programmable cleavage of target DNA.
- Ines Fonfara
- , Hagen Richter
- & Emmanuelle Charpentier
-
Article |
 The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA
The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA
Here the m1A modification is discovered in messenger RNA and mapped at the transcriptome-wide level; the modification is conserved, dynamic, accumulates in structured regions around translation initiation sites upstream of the first splice site, and correlates with higher protein expression.
- Dan Dominissini
- , Sigrid Nachtergaele
- & Chuan He
-
Article |
 Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution
Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution
A 3.7 Å resolution structure for the yeast U4/U6.U5 tri-snRNP, a complex involved in splicing, allows a better appreciation of the architecture of the tri-snRNP, and offers new functional insights into the activation of the spliceosome and the assembly of the catalytic core.
- Thi Hoang Duong Nguyen
- , Wojciech P. Galej
- & Kiyoshi Nagai
-
Letter |
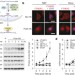 Dynamic m6A mRNA methylation directs translational control of heat shock response
Dynamic m6A mRNA methylation directs translational control of heat shock response
Under stress, such as heat shock, the N6-methyladenosine (m6A) modification is shown to accumulate primarily in the 5′ untranslated region of induced mRNAs owing to the translocation of an m6A interacting protein, YTHDF2, into the nucleus, resulting in increased cap-independent translation of these mRNAs, indicating one possible mechanism by which stress-responsive genes can be preferentially expressed.
- Jun Zhou
- , Ji Wan
- & Shu-Bing Qian
-
Article |
 Structure of mammalian eIF3 in the context of the 43S preinitiation complex
Structure of mammalian eIF3 in the context of the 43S preinitiation complex
The cryo-electron microscopy structure of the eukaryotic initiation factor 3 (eIF3) within the larger 43S complex is determined; the improved resolution enables visualization of the secondary structures of the subunits, as well as the contacts between eIF3 and both eIF2 and DHX29.
- Amedee des Georges
- , Vidya Dhote
- & Yaser Hashem
-
Article |
 RNA degradation paths in a 12-subunit nuclear exosome complex
RNA degradation paths in a 12-subunit nuclear exosome complex
Solving the crystal structure of an exosome complex from yeast, bound to different RNA substrates, offers insights into how the exosome can be utilized for precise processing of some 3′ ends, such as that of the 5.8S rRNA, while other RNAs are degraded to completion.
- Debora Lika Makino
- , Benjamin Schuch
- & Elena Conti
-
Letter |
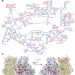 Protein synthesis by ribosomes with tethered subunits
Protein synthesis by ribosomes with tethered subunits
A ribosome with tethered subunits, ‘Ribo-T’, is engineered by making a hybrid RNA composed of ribosomal RNA of large and small subunits; Ribo-T can support cell growth in vivo in the absence of wild-type ribosomes, and is used to establish a fully orthogonal ribosome–mRNA system.
- Cédric Orelle
- , Erik D. Carlson
- & Alexander S. Mankin
-
Article |
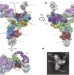 The architecture of the spliceosomal U4/U6.U5 tri-snRNP
The architecture of the spliceosomal U4/U6.U5 tri-snRNP
This study determines the structure of the spliceosomal tri-snRNP complex (containing three small nuclear RNAs and more than 30 proteins) by single-particle cryo-electron microscopy; the resolution is sufficient to discern the organization of RNA and protein components involved in spliceosome activation, exon alignment and catalysis.
- Thi Hoang Duong Nguyen
- , Wojciech P. Galej
- & Kiyoshi Nagai
-
Letter |
 Genome-wide identification of zero nucleotide recursive splicing in Drosophila
Genome-wide identification of zero nucleotide recursive splicing in Drosophila
In flies, some introns contain internal splice sites that cause ‘recursive splicing’, a multi-step removal of a single intron; this study demonstrates that the scope of this regulatory mechanism is much more extensive in flies than had been appreciated, and provides details about the recursive splicing process.
- Michael O. Duff
- , Sara Olson
- & Brenton R. Graveley
-
Letter |
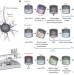 Synthesis and applications of RNAs with position-selective labelling and mosaic composition
Synthesis and applications of RNAs with position-selective labelling and mosaic composition
A hybrid solid–liquid phase transcription method and automated robotic platform synthesizes position-specific, fluorescence- or isotope-labelled RNA.
- Yu Liu
- , Erik Holmstrom
- & Yun-Xing Wang
-
Article |
 Structure of the human 80S ribosome
Structure of the human 80S ribosome
The structure of the human ribosome at high resolution has been solved; by combining single-particle cryo-EM and atomic model building, local resolution of 2.9 Å was achieved within the most stable areas of the structure.
- Heena Khatter
- , Alexander G. Myasnikov
- & Bruno P. Klaholz
-
Letter |
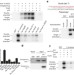 Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex
Defining fundamental steps in the assembly of the Drosophila RNAi enzyme complex
The assembly of single Drosophila RNA-induced silencing complexes (RISCs) is reconstituted using seven purified proteins, revealing that chaperones help stabilize the interaction of the protein heterodimer Dicer-2–R2D2 bound to the short interfering RNA with Ago2.
- Shintaro Iwasaki
- , Hiroshi M. Sasaki
- & Yukihide Tomari
-
Letter |
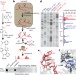 Structural imprints in vivo decode RNA regulatory mechanisms
Structural imprints in vivo decode RNA regulatory mechanisms
The single-stranded nature of RNAs synthesized in the cell gives them great scope to form different structures, but current methods to measure RNA structure in vivo are limited; now, a new methodology allows researchers to examine all four nucleotides in mouse embryonic stem cells.
- Robert C. Spitale
- , Ryan A. Flynn
- & Howard Y. Chang
-
Letter |
 hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1
hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1
A method, termed hiCLIP, has been developed to determine the RNA duplexes bound by RNA-binding proteins, revealing an unforeseen prevalence of long-range duplexes in 3′ untranslated regions (UTRs), and a decreased incidence of SNPs in duplex-forming regions; the results also show that RNA structure is able to regulate gene expression.
- Yoichiro Sugimoto
- , Alessandra Vigilante
- & Jernej Ule
-
Article |
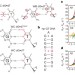 Visualizing transient Watson–Crick-like mispairs in DNA and RNA duplexes
Visualizing transient Watson–Crick-like mispairs in DNA and RNA duplexes
dG•dT and rG•rU ‘wobble’ mispairs in DNA and RNA transiently form base pairs with Watson–Crick geometry via tautomerization and ionization with probabilities that correlate with misincorporation probabilities during replication and translation.
- Isaac J. Kimsey
- , Katja Petzold
- & Hashim M. Al-Hashimi
-
Letter |
 N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions
N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions
The binding motifs for many RNA-binding proteins are normally buried within structured regions; now, the N6-methyladenosine modification is shown to act as a switch to remodel these regions, expose the motif, and thereby facilitate binding of RNA-binding proteins.
- Nian Liu
- , Qing Dai
- & Tao Pan
-
Letter |
 Initiation of translation in bacteria by a structured eukaryotic IRES RNA
Initiation of translation in bacteria by a structured eukaryotic IRES RNA
A eukaryotic viral internal ribosome entry site (IRES) element is described that binds both bacterial and eukaryotic ribosomes and initiates translation in both, demonstrating that RNA structure-based initiation can occur in both these domains of life, although in bacteria the element uses a mechanism that differs from that in eukaryotes.
- Timothy M. Colussi
- , David A. Costantino
- & Jeffrey S. Kieft
-
Letter |
 Catalysts from synthetic genetic polymers
Catalysts from synthetic genetic polymers
Four different XNAs — polymers with backbone chemistries not found in nature, namely, arabino nucleic acids, 2′-fluoroarabino nucleic acids, hexitol nucleic acids and cyclohexene nucleic acids — are found to be able to support the evolution of synthetic enzymes (XNAzymes) that catalyse several chemical reactions.
- Alexander I. Taylor
- , Vitor B. Pinheiro
- & Philipp Holliger
-
Letter |
 A cross-chiral RNA polymerase ribozyme
A cross-chiral RNA polymerase ribozyme
Here, a cross-chiral RNA polymerase is developed—an RNA enzyme that can catalyse the templated polymerization of activated mononucleotides that are of the opposite handedness—shedding light on how RNA-based life could have emerged.
- Jonathan T. Sczepanski
- & Gerald F. Joyce
-
Letter |
 R-loops induce repressive chromatin marks over mammalian gene terminators
R-loops induce repressive chromatin marks over mammalian gene terminators
R-loops, which have been considered to be rare and potentially harmful transcriptional by-products, are now shown to be needed for antisense transcription and to induce repressive chromatin marks that reinforce pausing of transcription and thereby enhance its termination.
- Konstantina Skourti-Stathaki
- , Kinga Kamieniarz-Gdula
- & Nicholas J. Proudfoot
-
Letter |
 The complete structure of the large subunit of the mammalian mitochondrial ribosome
The complete structure of the large subunit of the mammalian mitochondrial ribosome
The structure of the 39S large mitoribosome subunit is solved by cryo-electron microscopy at an impressive 3.4 Å resolution, revealing the location of 50 ribosomal proteins, the peptidyl transferase centre, the tRNAs within this active site, and the nascent peptide chain within the exit tunnel.
- Basil J. Greber
- , Daniel Boehringer
- & Nenad Ban
-
Letter |
 Programmable RNA recognition and cleavage by CRISPR/Cas9
Programmable RNA recognition and cleavage by CRISPR/Cas9
In the presence of a short DNA oligonucleotide containing a protospacer adjacent motif, a guide-RNA-programmed Cas9 is able to specifically bind and/or cleave single-stranded RNA—this system can be used to isolate specific endogenous RNA transcripts from a cell lysate without any tag or modification.
- Mitchell R. O’Connell
- , Benjamin L. Oakes
- & Jennifer A. Doudna
-
Article |
 Crystal structure of a eukaryotic group II intron lariat
Crystal structure of a eukaryotic group II intron lariat
This study determines the structure of a branched lariat RNA, providing insights into rearrangement of the intron between the two steps of RNA splicing.
- Aaron R. Robart
- , Russell T. Chan
- & Navtej Toor
-
Letter |
 Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
The CRISPR/Cas system is an RNA-guided bacterial protection system against foreign nucleic acids of bacterial and archaeal origin; here a high-resolution crystal structure of the CRIPSR RNA–Cas complex shows that the CRIPSR RNA plays an essential role not only in target recognition but also in complex assembly.
- Hongtu Zhao
- , Gang Sheng
- & Yanli Wang
-
Article |
 Structure of an Rrp6–RNA exosome complex bound to poly(A) RNA
Structure of an Rrp6–RNA exosome complex bound to poly(A) RNA
The exosome complex contains two catalytic subunits which degrade RNA in either a distributive (Rrp6) or a processive (Rrp44) manner—previous structures indicated how RNA could be directed to Rrp44, but the path taken to Rrp6 was unclear; here the location of the Rrp6 catalytic domain and the RNA 3′ end are determined and it is found that the RNA lies in an opposite orientation from that of the Rrp44-containing exosome structure, suggesting that the fate of an RNA may be influenced by the manner in which cofactors present it.
- Elizabeth V. Wasmuth
- , Kurt Januszyk
- & Christopher D. Lima
-
Letter |
 The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA
The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA
RNA molecules can perform multiple functions, which can be driven by different conformational states; here, the crystal structure of the transfer-RNA-like structure of the turnip yellow mosaic virus is solved, providing insight into the structural basis of RNA multifunctionality.
- Timothy M. Colussi
- , David A. Costantino
- & Jeffrey S. Kieft
-
Letter |
 Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors
Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors
Using a phylogenetic approach, the protein archease is identified as being a subunit of the human transfer RNA splicing ligase, and found to be necessary for full ligase activity, in cooperation with DDX1.
- Johannes Popow
- , Jennifer Jurkin
- & Javier Martinez
-
Article |
 Structural basis of the non-coding RNA RsmZ acting as a protein sponge
Structural basis of the non-coding RNA RsmZ acting as a protein sponge
A novel combined NMR and EPR spectroscopy approach reveals the structure and assembly mechanism of a 70-kDa bacterial ribonucleoprotein complex acting as a protein sponge in translational regulation.
- Olivier Duss
- , Erich Michel
- & Frédéric H.-T. Allain
-
Article |
 Protein-guided RNA dynamics during early ribosome assembly
Protein-guided RNA dynamics during early ribosome assembly
Three-colour fluorescence resonance energy transfer and molecular dynamics simulations are used to study the events occurring early in assembly of the 30S ribosome; within a non-native intermediate S4 ribosomal protein–16S RNA structure, S4 is capable of altering the RNA helix dynamics to facilitate conformation changes that enable subsequent protein binding.
- Hajin Kim
- , Sanjaya C. Abeysirigunawarden
- & Sarah A. Woodson
-
Article |
 Poly(A)-tail profiling reveals an embryonic switch in translational control
Poly(A)-tail profiling reveals an embryonic switch in translational control
A new high-throughput sequencing method to determine mRNA poly(A)-tail length enabled studies of individual RNAs across species and developmental stages to investigate the role of poly(A) length in translational regulation; the relationship between poly(A) length and translational efficiency shown in early embryo systems does not occur later in development, a finding that explains different regulatory consequences of microRNAs acting at different developmental times.
- Alexander O. Subtelny
- , Stephen W. Eichhorn
- & David P. Bartel
-
Article |
 DNA interrogation by the CRISPR RNA-guided endonuclease Cas9
DNA interrogation by the CRISPR RNA-guided endonuclease Cas9
This study defines how a short DNA sequence, known as the PAM, is critical for target DNA interrogation by the CRISPR-associated enzyme Cas9 — DNA melting and heteroduplex formation initiate near the PAM and extend directionally through the remaining target sequence, and the PAM is also required to activate the catalytic activity of Cas9.
- Samuel H. Sternberg
- , Sy Redding
- & Jennifer A. Doudna
-
Article |
 Architecture of the large subunit of the mammalian mitochondrial ribosome
Architecture of the large subunit of the mammalian mitochondrial ribosome
Cryo-electron microscopy combined with chemical crosslinking and mass spectrometry is used to determine the structure of the large subunit of the mammalian mitoribosome; this structure provides detailed structural insight, particularly of the molecular architecture of the polypeptide exit site, which has been structurally remodelled during evolution, presumably to help facilitate the membrane insertion of the highly hydrophobic proteins encoded by the mitochondrial genome.
- Basil J. Greber
- , Daniel Boehringer
- & Nenad Ban
-
Letter |
 Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo
Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo
Understanding how RNA structure influences its function has been hampered by a lack of approaches that can accurately quantify RNA structure in vivo; here, RNA structure is revealed on a global scale and with nucleotide-level resolution, showing that there is less structure within cells than expected from in vitro and in silico analyses.
- Silvi Rouskin
- , Meghan Zubradt
- & Jonathan S. Weissman
-
Letter |
 Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA
Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA
The crystal structure of the Lsm protein ring of the U6 small nuclear ribonucleoprotein (snRNP), with and without an RNA comprising the 3′ end of the U6 small nuclear RNA, is solved here; this structure provides insight into the function of U6 snRNP in precursor messenger RNA splicing.
- Lijun Zhou
- , Jing Hang
- & Yigong Shi
-
Letter |
 Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export
Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export
Two proteins are identified in yeast that regulate the timing of pre-ribosome export from the nucleus; Nug2 binds pre-60S particles until they are ready for export, at which time Nug2 is replaced by the export adaptor Nmd3, enabling the export machinery to recognise the pre-ribosome that is ready to be transferred to the cytoplasm.
- Yoshitaka Matsuo
- , Sander Granneman
- & Ed Hurt
-
Article |
 RNA catalyses nuclear pre-mRNA splicing
RNA catalyses nuclear pre-mRNA splicing
The spliceosome is shown to catalyse splicing through the RNA and not the protein components of the spliceosome; pre-messenger RNA splicing requires U6 snRNA acting by a mechanism similar to that used by group II self-splicing introns.
- Sebastian M. Fica
- , Nicole Tuttle
- & Joseph A. Piccirilli
-
Letter |
 Structural basis for the modular recognition of single-stranded RNA by PPR proteins
Structural basis for the modular recognition of single-stranded RNA by PPR proteins
Although the roles of pentatricopeptide repeat (PPR) proteins in RNA metabolism are well characterised, the mechanism by which they recognise specific single-stranded (ss)RNAs remains ill-understood; here X-ray crystal structures of maize PPR10 in the presence and absence of ssRNA provide details of the PPR10–ssRNA interaction.
- Ping Yin
- , Quanxiu Li
- & Nieng Yan
-
Letter |
 Hidden specificity in an apparently nonspecific RNA-binding protein
Hidden specificity in an apparently nonspecific RNA-binding protein
A novel high-throughput sequencing kinetics approach is used to measure functional binding of the apparently nonspecific RNA-binding protein C5 to all possible sequence variants in its substrate binding site; C5 binds different substrate variants with affinities varying widely, and with a similar affinity distribution to that of highly specific nucleic-acid-binding proteins, but it does not bind its physiological RNA targets with the highest affinity.
- Ulf-Peter Guenther
- , Lindsay E. Yandek
- & Eckhard Jankowsky
-
Letter |
 Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA
Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA
The co-crystal structure of the T-box tRNA-binding region, stem I, bound to tRNA is solved, showing that this region not only binds the anticodon, but also cradles the entire tRNA, forming an extended interface; the two T-loop motifs of stem I mediate interactions similar to those of RNase P and the large ribosomal subunit, even though the three species do not share a common evolutionary ancestor.
- Jinwei Zhang
- & Adrian R. Ferré-D’Amaré
-
Article |
 The initiation of mammalian protein synthesis and mRNA scanning mechanism
The initiation of mammalian protein synthesis and mRNA scanning mechanism
Three structures of the eukaryotic small ribosomal subunit in complex with initiator tRNA, mRNA and the initiation factors eIF1 and eIF1A have been solved; these structures offer insight into the contributions of the initiation factors, the mechanism by which mRNA is scanned, and the interactions that occur in the ribosome’s P site.
- Ivan B. Lomakin
- & Thomas A. Steitz
-
Article |
 A compendium of RNA-binding motifs for decoding gene regulation
A compendium of RNA-binding motifs for decoding gene regulation
This study reports a global analysis of binding sites for over 200 RNA-binding proteins (RBPs) from 24 species; conserved RNA-binding motifs are identified, and their analysis allows prediction of interaction sites based on the sequence of the RNA-binding domain alone.
- Debashish Ray
- , Hilal Kazan
- & Timothy R. Hughes
-
Letter |
 Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Members of the SAM-dependent methyltransferase superfamily are involved in the modification of wobble uridine to 5-oxacetyl uridine in Gram-negative bacteria; CmoA converts SAM to carboxy-SAM (Cx-SAM; a metabolite that was unknown previously), and CmoB uses Cx-SAM to convert 5-hydroxyuridine to 5-oxyacetyl uridine in tRNA.
- Jungwook Kim
- , Hui Xiao
- & Steven C. Almo
-
Article |
 Circular RNAs are a large class of animal RNAs with regulatory potency
Circular RNAs are a large class of animal RNAs with regulatory potency
Biochemical, functional and computational analyses are combined to show that circular RNAs are a large class of animal RNAs with regulatory potency.
- Sebastian Memczak
- , Marvin Jens
- & Nikolaus Rajewsky
-
Article |
 Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex
Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex
The crystal structure of a complete yeast exosome (Exo-10) bound to a region of the Rrp6 nuclease and an RNA substrate is determined, demonstrating that the exosome binds and degrades RNA molecules with a channelling mechanism that is largely conserved in all kingdoms of life and is similar to the mechanism used by the proteasome to degrade polypeptides.
- Debora Lika Makino
- , Marc Baumgärtner
- & Elena Conti
-
Research Highlights |
RNA tails time protein production
-
Article |
 FMRP targets distinct mRNA sequence elements to regulate protein expression
FMRP targets distinct mRNA sequence elements to regulate protein expression
RNA-recognition elements are identified for the fragile-X-syndrome-associated RNA-binding protein FMRP, in addition to its target messenger RNAs; although many of FMRP gene targets discovered are involved in brain function and autism spectrum disorder, a proportion are also dysregulated in mouse ovaries, suggesting cross-regulation of signalling pathways in different tissues.
- Manuel Ascano
- , Neelanjan Mukherjee
- & Thomas Tuschl
-
Letter |
 Activated GTPase movement on an RNA scaffold drives co-translational protein targeting
Activated GTPase movement on an RNA scaffold drives co-translational protein targeting
Single-molecule fluorescence microscopy techniques are used to elucidate features of the highly conserved protein-targeting machinery known as the signal recognition particle (SRP); the long SRP RNA is shown to be crucial for correct timing and precision of cargo handover to the protein-translocation machinery, a finding that could help to explain how other ribonucleosome complexes function during complex cellular processes.
- Kuang Shen
- , Sinan Arslan
- & Shu-ou Shan

 Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes