Article
|
Open Access
Featured
-
-
Article
| Open Access Mitotic clustering of pulverized chromosomes from micronuclei
Mitotic clustering of pulverized chromosomes from micronuclei
The CIP2A–TOPBP1 complex tethers fragmented chromosomes from micronuclei for asymmetric mitotic inheritance, explaining distinct patterns of chromosome rearrangements in cancers and genomic disorders.
- Yu-Fen Lin
- , Qing Hu
- & Peter Ly
-
Article
| Open Access Nuclear chromosome locations dictate segregation error frequencies
Nuclear chromosome locations dictate segregation error frequencies
Using single-cell DNA sequencing after an error-prone mitosis in untransformed, diploid cell lines and organoids, chromosomes are shown to have different segregation error frequencies that result in non-random aneuploidy landscapes.
- Sjoerd J. Klaasen
- , My Anh Truong
- & Geert J. P. L. Kops
-
Article |
 Structural basis of human separase regulation by securin and CDK1–cyclin B1
Structural basis of human separase regulation by securin and CDK1–cyclin B1
Structures of separase in complex with either securin or cyclin B–CDK1 shed light on the regulation of chromosome separation during the cell cycle.
- Jun Yu
- , Pierre Raia
- & Andreas Boland
-
Article |
 Centromeres are dismantled by foundational meiotic proteins Spo11 and Rec8
Centromeres are dismantled by foundational meiotic proteins Spo11 and Rec8
The meiotic proteins Spo11 and Rec8, which ensure meiotic recombination and reductional chromosome segregation, have additional activities that challenge centromere stability by promoting centromeric nucleosome remodelling in both fission yeast and human cells.
- Haitong Hou
- , Eftychia Kyriacou
- & Julia Promisel Cooper
-
Article |
 Regulation of the MLH1–MLH3 endonuclease in meiosis
Regulation of the MLH1–MLH3 endonuclease in meiosis
Reconstitution of the activation of the MLH1–MLH3 endonuclease shows how crossovers are formed during meiosis.
- Elda Cannavo
- , Aurore Sanchez
- & Petr Cejka
-
Article |
 Convergent genes shape budding yeast pericentromeres
Convergent genes shape budding yeast pericentromeres
The three-dimensional structure of pericentromeres in budding yeast is defined by convergent genes, which mark pericentromere borders and trap cohesin complexes loaded at centromeres, generating an architecture that allows correct chromosome segregation.
- Flora Paldi
- , Bonnie Alver
- & Adele L. Marston
-
Article |
 Securin-independent regulation of separase by checkpoint-induced shugoshin–MAD2
Securin-independent regulation of separase by checkpoint-induced shugoshin–MAD2
Shugoshin and MAD2 regulate separase-mediated chromosome separation during mitosis, in parallel to a previously identified mechanism involving the anaphase inhibitor securin.
- Susanne Hellmuth
- , Laura Gómez-H
- & Olaf Stemmann
-
Letter |
 Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome
Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome
Cryo-electron microscopy structures of the Saccharomyces cerevisiae inner kinetochore complex provide insights into the interdependencies of constituent subcomplexes and the mechanism of centromeric nucleosome recognition.
- Kaige Yan
- , Jing Yang
- & David Barford
-
Letter |
 DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome
DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome
In the absence of DHX9, circular RNAs accumulate and transcription and translation are dysregulated—effects that are exacerbated by concomitant depletion of the RNA-editing enzyme ADAR.
- Tuğçe Aktaş
- , İbrahim Avşar Ilık
- & Asifa Akhtar
-
Letter |
 Molecular mechanism for the regulation of yeast separase by securin
Molecular mechanism for the regulation of yeast separase by securin
The crystal structure of yeast separase in complex with its inhibitor securin sheds light on the mechanism of inhibition, in which securin inhibits separase by inserting a short segment into the active site.
- Shukun Luo
- & Liang Tong
-
Letter |
 Ki-67 acts as a biological surfactant to disperse mitotic chromosomes
Ki-67 acts as a biological surfactant to disperse mitotic chromosomes
During cell division, chromosomes are maintained as individual units; this process is shown to be mediated by the cell proliferation marker Ki-67, which has biophysical properties similar to those of surfactants.
- Sara Cuylen
- , Claudia Blaukopf
- & Daniel W. Gerlich
-
Letter |
 Structural basis of cohesin cleavage by separase
Structural basis of cohesin cleavage by separase
The crystal structures of the protease domain of separase are reported, showing how separase recognizes cohesin, and how phosphorylation of the cleavage site enhances separase activity.
- Zhonghui Lin
- , Xuelian Luo
- & Hongtao Yu
-
Letter |
 Replication stress activates DNA repair synthesis in mitosis
Replication stress activates DNA repair synthesis in mitosis
Common fragile sites (CFSs) are difficult-to-replicate regions of eukaryotic genomes that are sensitive to replication stress and that require resolution by the MUS81–EME1 endonuclease to re-initiate POLD3-dependent DNA synthesis in early mitosis; this study defines the specific pathway of events causing the CFS fragility phenotype.
- Sheroy Minocherhomji
- , Songmin Ying
- & Ian D. Hickson
-
Article |
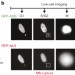 Chromothripsis from DNA damage in micronuclei
Chromothripsis from DNA damage in micronuclei
The mechanism for chromothripsis, “shattered” chromosomes that can be observed in cancer cells, is unknown; here, using live-cell imaging and single-cell sequencing, chromothripsis is shown to occur after a chromosome is isolated into a micronucleus, an abnormal nuclear structure.
- Cheng-Zhong Zhang
- , Alexander Spektor
- & David Pellman
-
Letter |
 Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation
Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation
Cyclin A is shown to maintain unstable kinetochore–microtubule (k–MT) attachments in prometaphase in order to allow for error correction; at the prometaphase–metaphase switch, k-MT attachments are stabilized when cyclin A drops below threshold levels.
- Lilian Kabeche
- & Duane A. Compton
-
Letter |
 BAF complexes facilitate decatenation of DNA by topoisomerase IIα
BAF complexes facilitate decatenation of DNA by topoisomerase IIα
Mutations in the subunits of BAF chromatin-remodelling complexes are frequently found in human cancer; here deletion of BAF subunits or expression of mutants of the ATPase subunit BRG1 attenuates genome-wide binding of topoisomerase IIα, resulting in tangled chromosomes, anaphase bridges and G2/M arrest.
- Emily C. Dykhuizen
- , Diana C. Hargreaves
- & Gerald R. Crabtree
-
Letter |
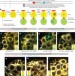 Chromosome-specific nonrandom sister chromatid segregation during stem-cell division
Chromosome-specific nonrandom sister chromatid segregation during stem-cell division
Using a CO-FISH method with single-chromosome resolution, sister chromatids of the sex chromosomes, but not autosomes, are shown to segregate nonrandomly during asymmetric cell divisions of Drosophila male germline stem cells; this suggests that it is unlikely that nonrandom sister chromatid segregation serves to protect the ‘immortal strand’ to avoid replication-induced mutations as proposed previously.
- Swathi Yadlapalli
- & Yukiko M. Yamashita
-
Letter |
 Tension sensing by Aurora B kinase is independent of survivin-based centromere localization
Tension sensing by Aurora B kinase is independent of survivin-based centromere localization
The current model to explain accurate chromosome segregation after DNA replication holds that kinetochore–microtubule attachments exert tension across the centromere and are stabilized by spatial separation from inner centromere-localized Aurora B; here an alternative model is presented, wherein active Aurora B produced by clustering is sufficient to ensure biorientation through a mechanism that is intrinsic to the kinetochore.
- Christopher S. Campbell
- & Arshad Desai
-

 Heritable transcriptional defects from aberrations of nuclear architecture
Heritable transcriptional defects from aberrations of nuclear architecture Towards building a chromosome segregation machine
Towards building a chromosome segregation machine