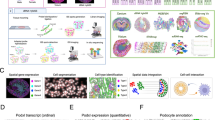
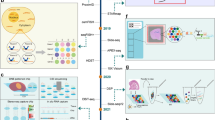
Abstract
The ability to localize hundreds of macromolecules to discrete locations, structures and cell types in a tissue is a powerful approach to understand the cellular and spatial organization of an organ. Spatially resolved transcriptomic technologies enable mapping of transcripts at single-cell or near single-cell resolution in a multiplex manner. The rapid development of spatial transcriptomic technologies has accelerated the pace of discovery in several fields, including nephrology. Its application to preclinical models and human samples has provided spatial information about new cell types discovered by single-cell sequencing and new insights into the cell–cell interactions within neighbourhoods, and has improved our understanding of the changes that occur in response to injury. Integration of spatial transcriptomic technologies with other omics methods, such as proteomics and spatial epigenetics, will further facilitate the generation of comprehensive molecular atlases, and provide insights into the dynamic relationships of molecular components in homeostasis and disease. This Review provides an overview of current and emerging spatial transcriptomic methods, their applications and remaining challenges for the field.
Key points
-
Spatially resolved transcriptomic technologies allow the spatial mapping of transcripts at single-cell or near single-cell resolution in a multiplex manner, and currently include sequencing-based technologies and imaging-based methodologies. Sequencing-based technologies include whole transcriptome-wide in situ capture and region of interest-based spatial RNA analysis platforms; imaging-based technologies include a variety of massively multiplexed in situ hybridization methodologies.
-
Key data outputs from spatial transcriptomic technologies include the anchoring of cell types and states derived from disaggregated single-cell technologies, determination of spatially variable gene expression, and the annotation of functional neighbourhoods.
-
Spatial technologies enable the localization of biologically relevant cellular interactions in the histopathological context and identification of disease-relevant cell signalling pathways associated with morphological and histopathological changes.
-
Spatial technologies are rapidly advancing to provide single-cell signatures at a whole transcriptome level. Further, these technologies are becoming increasingly multiplexed with orthogonal technologies such as spatial proteomics or epigenomics to define gene and protein regulatory patterns.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gall, J. G. The origin of in situ hybridization – a personal history. Methods 98, 4–9 (2016).
Tian, L., Chen, F. & Macosko, E. Z. The expanding vistas of spatial transcriptomics. Nat. Biotechnol. 41, 773–782 (2023).
Lopez, R. et al. DestVI identifies continuums of cell types in spatial transcriptomics data. Nat. Biotechnol. 40, 1360–1369 (2022).
Li, H. et al. A comprehensive benchmarking with practical guidelines for cellular deconvolution of spatial transcriptomics. Nat. Commun. 14, 1548 (2023).
Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 22, 627–644 (2021).
Dries, R. et al. Advances in spatial transcriptomic data analysis. Genome Res. 31, 1706–1718 (2021).
Jain, S. et al. Advances and prospects for the Human BioMolecular Atlas Program (HuBMAP). Nat. Cell Biol. 25, 1089–1100 (2023).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The Human Cell Atlas: from vision to reality. Nature 550, 451–453 (2017).
Rozenblatt-Rosen, O. et al. The Human Tumor Atlas Network: charting tumor transitions across space and time at single-cell resolution. Cell 181, 236–249 (2020).
de Boer, I. H. et al. Rationale and design of the Kidney Precision Medicine Project. Kidney Int. 99, 498–510 (2021).
HuBMAP Consortium. The human body at cellular resolution: the NIH human biomolecular atlas program. Nature 574, 187–192 (2019).
SenNet, C. NIH SenNet Consortium to map senescent cells throughout the human lifespan to understand physiological health. Nat. Aging 2, 1090–1100 (2022).
Oxburgh, L. et al. (Re)Building a Kidney. J. Am. Soc. Nephrol. 28, 1370–1378 (2017).
Regev, A. et al. The Human Cell Atlas. Elife 6, e27041 (2017).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792.e21 (2022).
Lake, B. B. et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature 619, 585–594 (2023).
Fan, Y. et al. Expansion spatial transcriptomics. Nat. Methods 20, 1179–1182 (2023).
Wirth, J. et al. Spatial transcriptomics using multiplexed deterministic barcoding in tissue. Nat. Commun. 14, 1523 (2023).
Salem, F. et al. The spatially resolved transcriptional profile of acute T cell-mediated rejection in a kidney allograft. Kidney Int. 101, 131–136 (2022).
Srivatsan, S. R. et al. Embryo-scale, single-cell spatial transcriptomics. Science 373, 111–117 (2021).
Lee, Y. et al. XYZeq: spatially resolved single-cell RNA sequencing reveals expression heterogeneity in the tumor microenvironment. Sci. Adv. 7, eabg4755 (2021).
Nguyen, H. Q. et al. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat. Methods 17, 822–832 (2020).
Lee, J. H. et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 10, 442–458 (2015).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Ke, R. et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 10, 857–860 (2013).
Groiss, S. et al. Highly resolved spatial transcriptomics for detection of rare events in cells. Preprint at bioRxiv, https://doi.org/10.1101/2021.10.11.463936 (2021).
Eng, C. L. et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019).
Andersson, A. et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 3, 565 (2020).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Melo Ferreira, R. et al. Integration of spatial and single cell transcriptomics localizes epithelial-immune cross-talk in kidney injury. JCI Insight 6, e147703 (2021).
Brbic, M. et al. Annotation of spatially resolved single-cell data with STELLAR. Nat. Methods 19, 1411–1418 (2022).
Wei, R. et al. Spatial charting of single-cell transcriptomes in tissues. Nat. Biotechnol. 40, 1190–1199 (2022).
Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I. & Heyn, H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 49, e50 (2021).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022).
Li, B. et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods 19, 662–670 (2022).
Melo Ferreira, R., Freije, B. J. & Eadon, M. T. Deconvolution tactics and normalization in renal spatial transcriptomics. Front. Physiol. 12, 812947 (2021).
Zeng, Z., Li, Y., Li, Y. & Luo, Y. Statistical and machine learning methods for spatially resolved transcriptomics data analysis. Genome Biol. 23, 83 (2022).
Qian, J. et al. Reconstruction of the cell pseudo-space from single-cell RNA sequencing data with scSpace. Nat. Commun. 14, 2484 (2023).
Wang, J. et al. Dimension-agnostic and granularity-based spatially variable gene identification using BSP. Nat. Commun. 14, 7367 (2023).
Keller, M. S. et al. Vitessce: a framework for integrative visualization of multimodal and spatially-resolved single-cell data. Preprint at OSF Preprints, https://doi.org/10.31219/osf.io/y8thv (2023).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Winfree, S. et al. Integrated cytometry with machine learning applied to high-content imaging of human kidney tissue for in situ cell classification and neighborhood analysis. Lab. Invest. 103, 100104 (2023).
Li, Z., Song, T., Yong, J. & Kuang, R. Imputation of spatially-resolved transcriptomes by graph-regularized tensor completion. PLoS Comput. Biol. 17, e1008218 (2021).
Park, J. et al. Cell segmentation-free inference of cell types from in situ transcriptomics data. Nat. Commun. 12, 3545 (2021).
Zhang, Y. et al. Reference-based cell type matching of in situ image-based spatial transcriptomics data on primary visual cortex of mouse brain. Sci. Rep. 13, 9567 (2023).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Canela, V. H. et al. A spatially anchored transcriptomic atlas of the human kidney papilla identifies significant immune injury in patients with stone disease. Nat. Commun. 14, 4140 (2023).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Llorens-Bobadilla, E. et al. Solid-phase capture and profiling of open chromatin by spatial ATAC. Nat. Biotechnol. 41, 1085–1088 (2023).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Gisch, D. L. et al. The chromatin landscape of healthy and injured cell types in the human kidney. Nat. Commun. 15, 433 (2024).
Lucarelli, N. et al. Correlating deep learning-based automated reference kidney histomorphometry with patient demographics and creatinine. Kidney360 4, 1726–1737 (2023).
Shickel, B. et al. Spatially aware transformer networks for contextual prediction of diabetic nephropathy progression from whole slide images. Proc. SPIE Int. Soc. Opt. Eng. 12471, 124710K (2023).
Zheng, Y. et al. Deep-learning-driven quantification of interstitial fibrosis in digitized kidney biopsies. Am. J. Pathol. 191, 1442–1453 (2021).
FUSION: Functional Unit State Identification and Navigation with WSI. Welcome to FUSION! fusion, http://fusion.hubmapconsortium.org/ (2023).
Ferkowicz, M. J. et al. Molecular signatures of glomerular neovascularization in a patient with diabetic kidney disease. Clin. J. Am. Soc. Nephrol. 19, 266–275 (2023).
Janosevic, D. et al. The orchestrated cellular and molecular responses of the kidney to endotoxin define a precise sepsis timeline. Elife 10, e62270 (2021).
Cheung, M. D. et al. Resident macrophage subpopulations occupy distinct microenvironments in the kidney. JCI Insight 7, e161078 (2022).
Wang, Z. et al. Integrated single-nucleus sequencing and spatial architecture analysis identified distinct injured-proximal tubular types in calculi rats. Cell Biosci. 13, 92 (2023).
Dixon, E. E., Wu, H., Muto, Y., Wilson, P. C. & Humphreys, B. D. Spatially resolved transcriptomic analysis of acute kidney injury in a female murine model. J. Am. Soc. Nephrol. 33, 279–289 (2022).
Wu, H. et al. High resolution spatial profiling of kidney injury and repair using RNA hybridization-based in situ sequencing. Nat. Commun. 15, 1396 (2024).
Kayhan, M. et al. Intrinsic TGF-β signaling attenuates proximal tubule mitochondrial injury and inflammation in chronic kidney disease. Nat. Commun. 14, 3236 (2023).
Onoda, N. et al. Spatial and single-cell transcriptome analysis reveals changes in gene expression in response to drug perturbation in rat kidney. DNA Res 29, dsac007 (2022).
Sanchez-Ferras, O. et al. A coordinated progression of progenitor cell states initiates urinary tract development. Nat. Commun. 12, 2627 (2021).
Hodgin, J. B. et al. Quantification of glomerular structural lesions: associations with clinical outcomes and transcriptomic profiles in nephrotic syndrome. Am. J. Kidney Dis. 79, 807–819.e1 (2022).
Mariani, L. H. et al. CureGN study rationale, design, and methods: establishing a large prospective observational study of glomerular disease. Am. J. Kidney Dis. 73, 218–229 (2019).
Townsend, R. R. et al. Rationale and design of the Transformative Research in Diabetic Nephropathy (TRIDENT) Study. Kidney Int. 97, 10–13 (2020).
Hickey, J. W. et al. Organization of the human intestine at single-cell resolution. Nature 619, 572–584 (2023).
Hunter, M. V., Moncada, R., Weiss, J. M., Yanai, I. & White, R. M. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat. Commun. 12, 6278 (2021).
Lake, B. B. et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 10, 2832 (2019).
Chen, D. et al. Single-cell RNA-seq with spatial transcriptomics to create an atlas of human diabetic kidney disease. Faseb J. 37, e22938 (2023).
Abedini, A. et al. Spatially resolved human kidney multi-omics single cell atlas highlights the key role of the fibrotic microenvironment in kidney disease progression. Preprint at bioRxiv, https://doi.org/10.1101/2022.10.24.513598 (2024).
Marshall, J. L. et al. High-resolution Slide-seqV2 spatial transcriptomics enables discovery of disease-specific cell neighborhoods and pathways. iScience 25, 104097 (2022).
Raghubar, A. M. et al. Spatially resolved transcriptomes of mammalian kidneys illustrate the molecular complexity and interactions of functional nephron segments. Front. Med. 9, 873923 (2022).
Cheung, M. D. et al. Spatiotemporal immune atlas of a clinical-grade gene-edited pig-to-human kidney xenotransplant. Nat. Commun. 15, 3140 (2024).
Zimmerman, S. M. et al. Spatially resolved whole transcriptome profiling in human and mouse tissue using digital spatial profiling. Genome Res. 32, 1892–1905 (2022).
El-Achkar, T. M. et al. A multimodal and integrated approach to interrogate human kidney biopsies with rigor and reproducibility: guidelines from the Kidney Precision Medicine Project. Physiol. Genomics 53, 1–11 (2021).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Long, Y. et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat. Commun. 14, 1155 (2023).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Fischer, D. S., Schaar, A. C. & Theis, F. J. Modeling intercellular communication in tissues using spatial graphs of cells. Nat. Biotechnol. 41, 332–336 (2023).
Acknowledgements
S.J. is supported by NIH funding (U54DK134301, U01DK114933, UH3DK114933 and P50DK133943). M.T.E. is supported by NIH funding (R01AT011463-01A1, U54 DK134301-01 and U01 DK114923).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Nephrology thanks Andrew Mallett, Christoph Kuppe and Yutaka Suzuki for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jain, S., Eadon, M.T. Spatial transcriptomics in health and disease. Nat Rev Nephrol (2024). https://doi.org/10.1038/s41581-024-00841-1
Accepted:
Published:
DOI: https://doi.org/10.1038/s41581-024-00841-1